Integrated In silico Docking and MoMA Simulation Approaches Reveal Withaferin A, Withalongolides A and B as Potent AldoKeto Reductase (AKR) 1C3 Inhibitors
Vadapalli J, Gallagher R, Vanam A, Motohashi N and Gollapudi R*
DOI10.21767/2469-6692.10026
Vadapalli J1, Gallagher R2, Vanam A3, Motohashi N4 and Gollapudi R2*
1Acharya Nagarjuna University, Nagarjunanagar, AP, India
2University of Kansas, Lawrence, KS, USA
3Sri Venkateswara University, Tirupathi, AP, India
4Meiji Pharmaceutical University, Noshio, Kiyose-shi, Tokyo, Japan
- *Corresponding Author:
- Rao Gollapudi
University of Kansas
Lawrence, KS 66045, USA
Tel: (785) 393 8872
E-mail: gollapudirao@ku.edu
Received Date: October 22, 2018; Accepted Date: December 13, 2018; Published Date: December 20, 2018
Citation: Vadapalli J, Gallagher R, Vanam A, Motohashi N, Gollapudi R (2018) Integrated In silico Docking and MoMA Simulation Approaches Reveal Withaferin A, Withalongolides A and B as Potent Aldo- Keto Reductase (AKR) 1C3 Inhibitors. J In Silico In Vitro Pharmacol 4:4. DOI: 10.21767/2469-6692.10026.
Abstract
Aldo-keto reductase family 1 member C3 (AKR1C3) is a steroidogenic enzyme and prospective drug target in leukemia, breast and prostate cancers. Overexpression of AKR1C3 can lead to the development of a variety of human cancers. Cancers that overexpress AKR1C3 exhibit chemotherapeutic resistance resulting in treatment failure and disease progression. Inhibition of AKR1C3 expression down-regulates cell proliferation in abiraterone-resistant prostate cancer cells. Therefore, the identification of a selective AKR1C3 inhibitor may reveal a novel strategy to combat drug resistance cancers. Recently, withaferin A (1) and other withanolides attracted attention as promising therapeutic candidates useful in treating a variety of cancers. Bioinformatic tools were utilized to identify the specific targets of different ligands. In silico predictions, modelling and dynamic simulations based on the crystal structure of AKR1C3 and ligands were performed. Active molecular docking studies using AutoDock Vina software suggested that withaferin A, withalongolide A and withalongolide B are novel inhibitors of AKR1C3 with binding affinities of -11.2, -12.5 and -13.1 kcal/mol, respectively. Of the ligands investigated in this study, withalongolide B produced the greatest AKR1C3 binding affinity. The data suggests that withalongolide B could be utilized as potent lead compound to probe cancers that utilize AKR1C3 overexpression for drug- resistance, such as abiraterone-resistant prostate cancer.
Keywords
Aldo-keto reductase family 1 member C3 (AKR1C3); Lipinski’s rule of five; Docking studies; In silico; Withaferin A; Withalongolide A; Withalongolide B; Withanolides
Abbreviations
AKR1C3: Aldo-Keto Reductase Family 1 member C3; RO5: Lipinski's Rule of Five
Introduction
Aldo-keto reductase family 1 member C3 (AKR1C3) enzyme, also known a17β-hydroxysteroid dehydrogenase type 5 (17β-HSD5, HSD17B5), hydroxysteroid (17-beta) dehydrogenase 5, and 3-alpha hydroxysteroid dehydrogenase, type II, is a strategic steroidogenic enzyme in humans that is encoded by the AKR1C3 gene located on chromosome 10 [1,2]. The AKR1C3 enzyme is essential in the biosynthesis of androgens and is considered an attractive target for the treatment of prostate cancer. AKR1C3 initiates to abiraterone resistance by increasing intracrine androgen synthesis, thereby improving the androgen signaling that occurs after androgen receptor (AR) activation. Overexpression of AKR1C3 offers resistance to abiraterone, whereas the down-regulation of AKR1C3 allows abiraterone-resistant cells to respond to abiraterone treatment. In abiraterone-resistant prostate cancer cells, intracrine androgen levels are elevated and AKR1C3 is overexpressed. Therefore, treatment of abiraterone-resistant cells with AKR1C3 inhibitors results in a decrease of intracrine androgen levels and reduces AR transcriptional activity, thereby incapacitating resistance and increasing the efficiency of abiraterone therapy [3]. The AKR1C3 overexpression in prostate and breast cancers occurs with the alteration in mRNA and/or protein expression levels within tumor tissues. Overexpression of AKR1C3 in LNCaP prostate cancer cells produced elevated testosterone production. Single-nucleotide polymorphisms in AKR1C3 are linked to disease progression and aggressiveness in prostate carcinomas. Studies have shown that AKR1C3 can induce resistance to a variety of anticancer drugs, such as doxorubicin, oracin and cisplatin. Furthermore, elevated AKR1C3 protein levels were associated with radioresistance in non-small cell lung carcinomas [4]. Collectively, AKR1C3 inhibition prevents cancer cell proliferation and thus offers an attractive target for the treatment of prostate cancer.
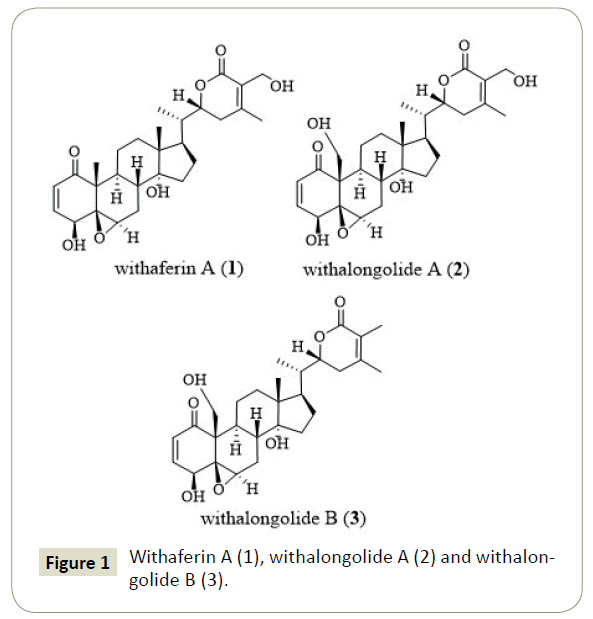
Recently, withaferin A (1) attracted attention as a promising therapeutic candidate helpful in treating a variety of cancers [5,6]. Withaferin A alone, or in combination with other treatments, was very effective as a novel alternative therapy against glioblastoma (GBM) [7,8]. In vitro and In vivo studies on withaferin A (1) and its 19-hydroxy derivatives (2 and 3) suggested that withalongolide A (2) and withalongolide B (3) curbed cellular growth via induction of apoptosis in a wide range of tumor cells including leukemia; breast, lung, pancreatic, and prostate cancers; as well as head and neck squamous cell carcinoma (HNSCC). In addition, withalongolides A (2) and B (3) displayed cytotoxicity against the HNSCC (JMAR, MDA-1986), melanoma (B16F10 and SKMEL-28), and/or normal fetal fibroblast (MRC-5) cells [9]. Withaferin A (1) analogues were explored for their anticancer potential and anti-proliferative activity as well as the underlying mechanisms in inhibiting the heat shock protein 90 (Hsp90) chaperone [10]. Withaferin A (1) reversed tumor progression by inducing a dose-dependent G2/M cell cycle arrest and promoting cell differentiation and death via the intrinsic and extrinsic apoptotic pathways [8]. In human breast cancer cells, withaferin A (1) attenuated estrogen receptor-α (ER-r) expression, which was attributed to the suppression of level of ER-v mRNA [11]. Several in silico studies on withaferin A (1) using various biological targets was reported [11-16]. Until now, no study has explored the potential of withaferin A (1) or any of the other withanolides as effective agents at controlling the AKR1C3 overexpression observed in cancer cells.
Molecular docking is a computer-assisted drug design that simulates the docking of a ligand into the active site of the receptor. The purpose of receptor and ligand docking interactions can be predicted through three-dimensional visualization of the docked complexes. This research was conducted in order to understand the binding interactions between receptor protein and the three naturally occurring withanolides, specifically withaferin A (1), withalongolide A (2) and withalongolide B (3) (Figure 1).
Materials and Methods
Lipinski’s rule of 5
In drug discovery projects, determination of the drug-likeness of a lead molecule is vital in order to reduce the experimental cost. The best lead candidate should not breach more than one of the criteria as defined by the “Lipinski's rule of five” (RO5). These rules include the optimum range of molecular weight (no greater than 500 Daltons or g/mol), octanol-water partition coefficient (Logp of 5 or less), hydrogen bond donor (5 or less) and hydrogen bond acceptor (10 or less) values that an ideal drug should contain. The drug likeliness tool (DruLiTo) software, a freely available software package, was used to determine the physicochemical characteristics of the three withanolides (1-3), namely withaferin A (1), withalongolide A (2), and withalongolide B (3) (Table 1). This analysis revealed that all three compounds satisfied the majority of the Lipinski criteria (Table 1) [17].
| Ligand | Mol. Wt (≤ 500) | Logp (≤ 5) | H-Donors (≤ 5) | H-Acceptors (≤ 10) |
|---|---|---|---|---|
| Withaferin A (1) | 431.97 | 3.987 | 6 | 0 |
| Withalongolide A (2) | 447.96 | 3.284 | 7 | 0 |
| Withalongolide B (3) | 431.97 | 3.892 | 6 | 0 |
Table 1: Examination of the “Lipinski’s rule of five” parameters in the three withanolides: withaferin A (1), withalongolide A (2) and withalongolide B (3).
Protein preparation
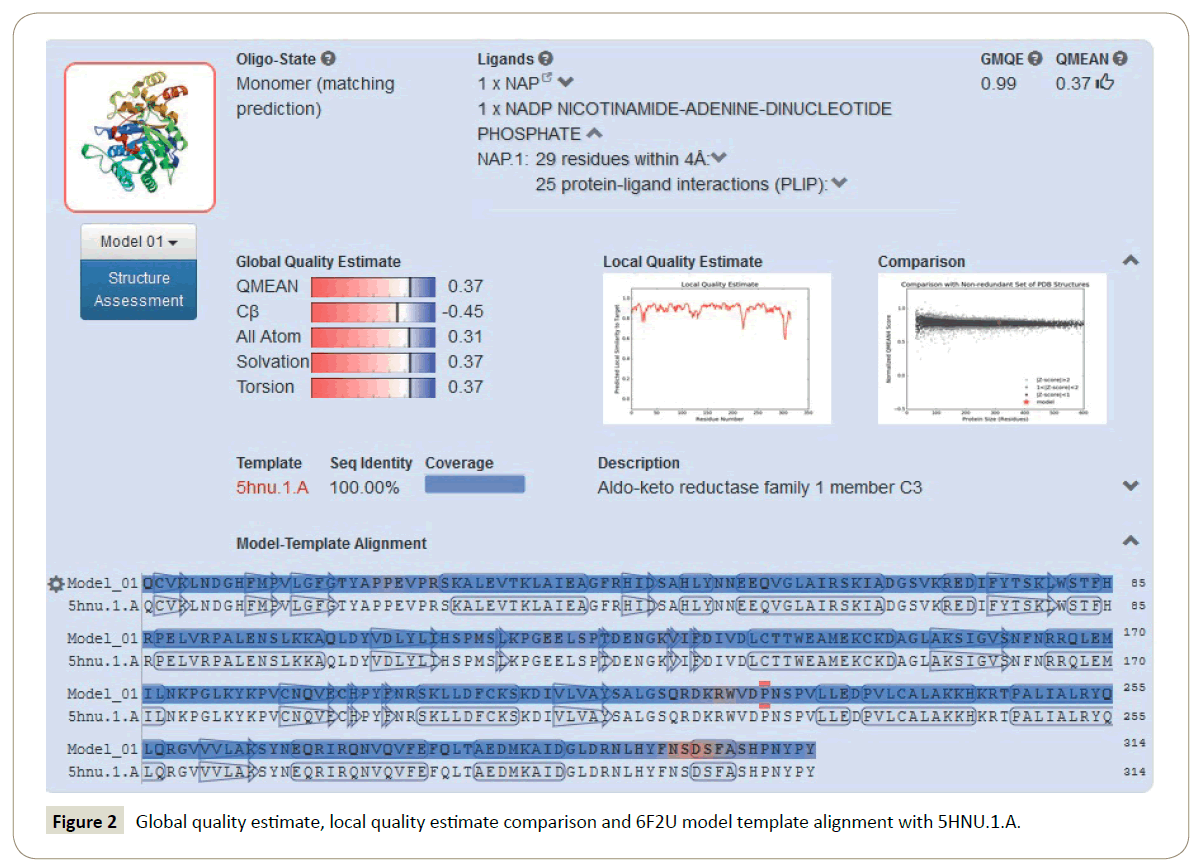
Homology modelling of the 6F2U protein: Homology modelling methods make use of experimental protein structures ("templates") to build receptor proteins in drug discovery. Homology (or comparative) modelling is currently the most accurate method to generate reliable three-dimensional protein structure models. SWISS-MODEL is a structural bioinformatics web-server dedicated to homology modelling to predict protein 3D structures [18,19]. The aldo-keto reductase, AKR1C3 (PDB ID: 6F2U), protein structure retrieved from protein data bank (https://www.rcsb.org/) was missing some amino acid residues. The complete AKR1C3 protein was modelled by submitting FASTA sequence of 6F2U (chain A) protein into SWISS-MODEL Workspace through automated mode for the development of a more accurate protein model [20]. The 6F2U (Chain A) protein and its sequence were selected as the target protein and query sequence, respectively. The crystal structure of AKR1C3 - octyl gallate complex (PDB ID: 5HNU) was also obtained from the protein data bank. The 6F2U protein model was built by using the 5HNU protein as a suitable template. In the SWISS MODEL Workspace automated mode, forty-four templates of query sequence (DszB protein sequence) were generated. The template, 5HNU.1.A, demonstrated highest sequence identity to query sequence and was used to develop an improved model of the 6F2U protein. Global quality estimate, local quality estimate comparison and 6F2U model template alignment with 5HNU.1.A were calculated (Figure 2).
Modeled AKR1C3 protein validation: Homology modeling methods utilize experimental protein structures ("templates") to build receptor proteins in drug discovery. Homology (or comparative) modeling is currently the most accurate method to generate reliable three-dimensional protein structure models. SWISS-MODEL is a structural bioinformatics web-server dedicated to homology modelling to predict protein 3D structures [21,22]. The structure has some missing amino acid residues. The AKR1C3 protein was modelled by submitting the FASTA sequence of 6F2U protein to the SWISS-MODEL Workspace through automated mode for the development of an accurate protein model [20].
The 6F2U protein and its sequence were selected as the target protein and query sequence, respectively. The protein model was built by using the 5HNU protein as a suitable template.
The 6F2U protein quality was validated by Ramachandran plot using Rampage [23] and in SPDBV (Deep View – Swiss – Pdb Viewer) version 4.10 based on the RMSD value obtained by superimposing the 6F2U protein model on its model template 5HNU [24]. The crystal structure of 6F2U-Modelled was selected for molecular docking to get reliable prediction of the ligands ability to bind with the receptor. The quality of the 6F2U-Modelled protein was validated by Ramachandran plot using Rampage [23] and in SPDBV (Deep View – Swiss – Pdb Viewer) version 4.10 based on the RMSD value obtained by superimposing 6F2U protein model on its model [24]. Ramachandran plot values of the 6F2U, 6F2U-Modelled protein and its template 5HNU.1.A were obtained (Table 2). In the Ramachandran plot generated for the 6F2U protein, 97.4% of the amino acid residues were found in favored region, 2.6% residues in allowed area, and 0% of the residues were present in the outlier regions (Figure 3). The Ramachandran plot of 5HNU.1.A showed 97.9% residues in favored region, 1.4% residues in allowed region, and 0.6% of the residues in the outlier regions. The Ramachandran plot generated for the 6F2U-Modelled protein displayed 98.1%, 1.9% and 0% of the residues in the favored, the allowed, and the outlier regions, respectively. The Ramachandran plot data for the 6F2U-Modelled, 6FTU and 5HNU proteins suggested favorable reliability of the 6F2U-Modelled protein for subsequent docking studies (Table 2) (Figure 3).
| Percent (%) of residues per region | |||
|---|---|---|---|
| Structure | favored | allowed | outlier |
| 6F2U-Modelled | 98.1 | 1.9 | 0 |
| 6F2U | 97.4 | 2.6 | 0 |
| 5HNU | 97.9 | 1.4 | 0.6 |
Table 2: The data from Ramachandran plots of the 6F2U-Modelled, 6F2U and 5HNU proteins.
Energy minimization and refinement of the modeled 6F2U protein (6F2U-Modelled): The modelled 6F2U (6F2U-Modelled) protein valency and chemistry were rectified using Chimera (UCSF, San Francisco, CA, USA). Energy minimization and refinement were performed by employing CHARMm force field in order to obtain a protein with least energy [25,26]. Later, polar hydrogen atoms were added to the protein model using the AutoDock tools 4.2.6 screening tool and PyRx v0.8 software (https://pyrx.sourceforge.net/).
Receptor cavity prediction: The cavity or the potential ligand binding site of AKR1C3 (PDB ID:6F2U) was predicted using MVD with the volume of cal-mol 358.4 A0 which was used for docking.
Preparation of ligands
The structures of withaferin A (1; CID: 265237), withalongolides A (2; CID: 56649343) and B (3; CID: 56649344) were initially retrieved from the PubChem Compound Database (National Center for Biotechnology Information, U.S. National Library of Medicine). Molecular geometry optimization of the ligands was achieved using Avogadro (an open-source molecular builder and visualization tool. Version 1.90.0. https://avogadro.cc/). The force field MMF94 was set with number of steps 500, algorithm steepest descent and convergence of 10e-7 [27]. The structures of ligands were saved in Protein Data Bank (PDB) file format and used for the docking study.
Docking between AKR1C3 and ligands
Discovery Studio visualizer and Chimera (UCSF, San Francisco, CA, USA) were chosen for visual inspection and preparations. PyRx software was employed as the virtual screening software. PyRx includes AutoDock Vina with a Lamarckian genetic algorithm as a scoring algorithm. The ligand/protein simulated interactions of withaferin A (1), withalongolides A (2) and B (3) with AKR1C3 were determined using AutoDock Vina (Molecular Graphics Lab, La Jolla, CA, USA) [27,28]. The docking was conducted with exhaustiveness of 8 and a grid box with the dimensions for center: 25×25×25 Å and box center: center_x=3.7981, center_y=-9.8707 center_z=2.6182 for 6F2U docking. PyMol v1.3 (Schrodinger, New York, NY, USA). AutoDock Vina evaluated target conformation (biomacromolecule) as a rigid unit while ligands were conceded to be flexible and adoptable to the target. The software determined the lowest binding affinity by using different conformations of each ligand. AutoDock Vina searched for the lowest binding affinity conformations and determined 9 different conformations for each receptor and ligand complexes. Each receptor and ligands complex with the lowest binding energy docking poses were selected. The protein-ligand interactions were analyzed with LigPlot and Discovery Studio 4.5 (Dassault Systemes BIOVIA, Discovery Studio Modelling Environment, Release 2017, San Diego, USA). Docking simulations of target AKR1C3 with withaferin A (1), withalongolide A (2) and withalongolide B (3) were performed in AutoDock Vina. (Molecular Graphics Lab, La Jolla, CA, USA). The AutoDock Vina software prepared the target in a rigid conformation while ligands were permitted to be flexible and malleable to the target. After the completion of the docking, ligand conformations displaying greatest binding affinity and lowest docked energies to the target were selected. The hydrogen bonds, bond lengths and hydrophobic interactions between AKR1C3 and ligands withaferin A (1), withalongolides A (2) and B (3) were determined by using LigPlot (https://www.ebi.ac.uk/thornton-srv/software/LIGPLOT/).
Target-ligand complex unbinding simulations
The MoMA-LigPath web server and Molecular Motion Algorithms (MoMA) were utilized to simulate the ligand unbinding from the binding site to surface of the target. In addition, the server discloses flexibility of protein side-chains, ligands and includes only statistical limitations. This process generates mechanistic data on the pathway of each ligand as it moves from the protein surface to the binding site or from binding site to surface of the protein. The program offers molecular interaction graphics, leading the ligands from surface of protein to the binding site. In this process, the program identifies certain residues that are crucial factors for ligand binding or driving ligands towards the binding site, in spite of being away from the binding site. The docked molecular complexes of AKR1C3 with withaferin A (1), withalongolides A (2) and B (3) that produced the lowest binding energies were selected for unbinding simulations by using MoMA LigPath [29,30].
Result and Discussion
In the Western world, prostate cancer is the most diagnosed cancer and the second leading cause of mortality in men. Hence, there is an immediate requirement for the discovery of novel agents that are capable of inhibiting the critical features of the resistance process that emerge after the androgen deprivation therapy (ADT). Overexpression of AKR1C3 is a critical mechanism of drug resistance observed in castration resistant prostate cancer cells. Hence, the development of potent AKR1C3-selective inhibitors is considered an important strategy in the treatment of prostate cancer and metastatic diseases [31,32].
The interpretation of the protein-ligand interactions plays an important role in structural based drug discovery. When each ligand (1-3) was docked with the protein receptor (ALR1C3) different binding energies were observed. Of these, withaferin A (1) produced the lowest value (-11.2 kcal/mol), followed by withalongolide A (2) (-12.5 kcal/mol), whereas withalongolide B (3) generated the greatest binding energy (-13.1 kcal/mol) when docked with the AKR1C3 receptor. These results suggest that these withanolides might emerge as promising candidates for the inhibition of AKR1C3 enzyme activity.
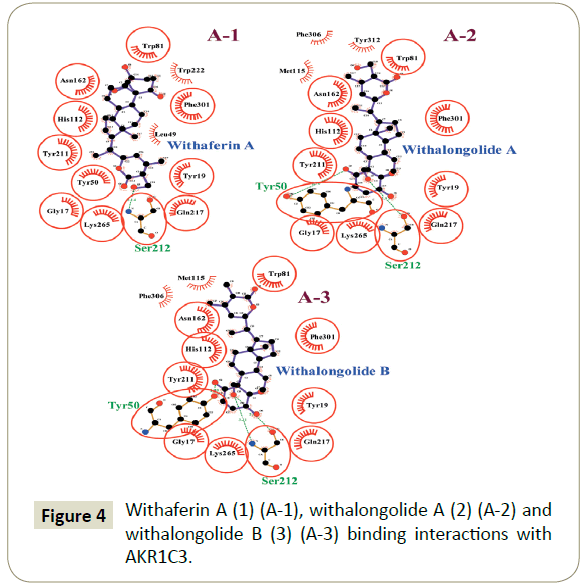
The present docking study explored the interactions of withaferin A (1), withalongolide A (2) and withalongolide B (3) with the receptor protein AKR1C3 (6F2U) and their binding patterns with AKR1C3 amino acid residues. Withaferin A (1), withalongolide A (2) and withalongolide B (3) docked with the active site of AKR1C3. Each amino acid residue within a 4A0 distance from the ligand was evaluated for the presence of any alkyl interaction, hydrogen bond, Pi-Sigma interaction, or van der Waals force. As a result, withaferin A (1) produced 10 van der Waals forces (Gly 17, Ser 113, Asn 162, Ser 212, Ala 213, Ser 216, Gln 217, Trp 222, Tys 265 and Phe 306) and 7 alkyl interactions (Tyr 19, Leu 49, Tyr 50, Trp 81, His 112, Tyr 211 and Phe 301) with AKR1C3; while withalongolide A (2) produced 13 van der Waals forces (Gly 17, Asp 45, Leu 49, Ser 113, Gln 185, Ser 212, Ser 216, Gln 217, Leu 263, Lys 265, Tyr 312, Pro 313 and Tyr 314), 5 alkyl interactions (Tyr19, Trp 81, His 112, Met 115 and Phe 396), and 3 hydrogen bonds (Tyr 50, Asn 162 and Tyr 211) ) with AKR1C3; whereas withalongolide B (3) generated 12 van der Waals forces (Gly 17, Asp 45, Leu 49, Lys 79, Ser 113, Gln 185, Ser 216, Gln 217, Leu 263, Lys 265, Tyr 312 and Tyr 314), 5 alkyl interactions (Tyr 19, Trp 81, His 112, Met 115 and Phe 306), 4 hydrogen bonds (Tyr 50, Asn 162, Tyr 211 and Ser 212) and a Pi-Sigma interaction (Phe 301) with AKR1C3 residues.
Combined, these results suggested among the withanolides investigated that withalongolide B (3) might function as the better AKR1C3 inhibitor as it produced the highest binding energy (-13.1 Kcal) (Figure 4).
Unbinding simulation of AKR1C3/withaferin A complex
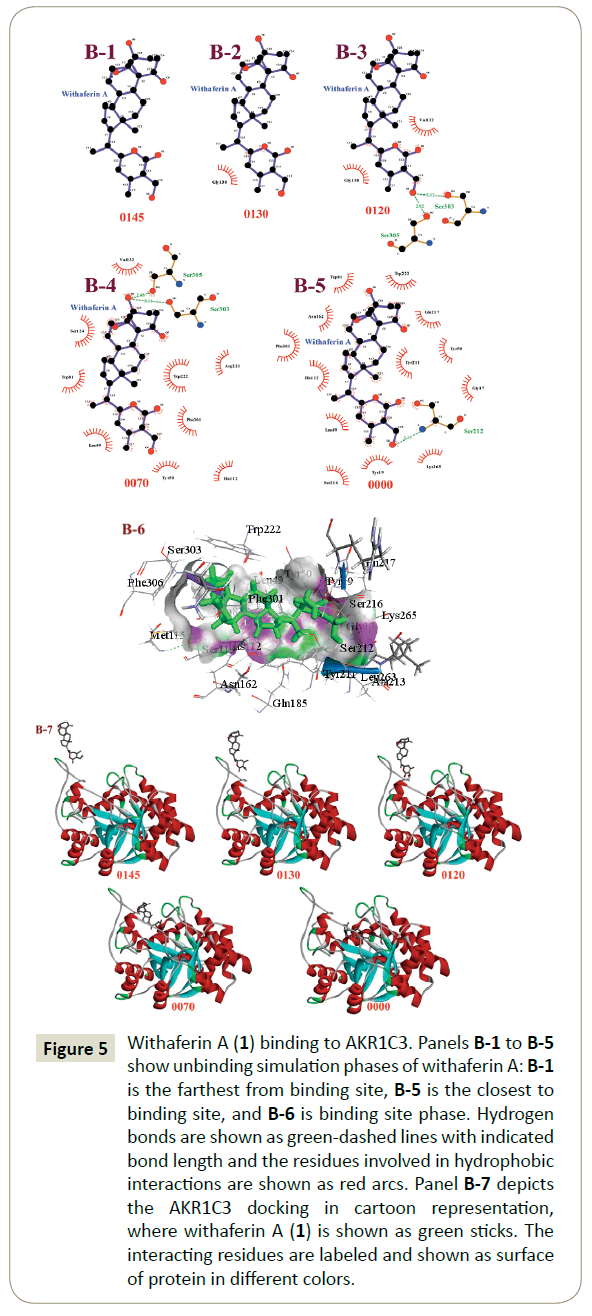
The unbinding simulation phases of withaferin A (1) with increasing number of molecular interactions while progressively approaching towards the binding site of AKR1C3 are shown in Figure 5 (B-1 to B-5). The docking phase displays the molecular interactions of AKR1C3 with withaferin A (1), bound on the exterior region of AKR1C3 cavity by hydrophobic interactions. In addition, withaferin A (1) was involved in interaction through Gly 130. The amino acid, Gly 130 was common in all phases of the unbinding simulation of withaferin A (1), which emphasized the importance of Gly 130 in stabilizing AKR1C3- withaferin A complex. A graphical representation of AKR1C3 in complex with withaferin A (1) was generated ((Figure 5) B-6) and a cartoon representation of the dynamic simulations that display the movement and binding of withaferin A with AKR1C3 to form the AKR1C3-withaferin A complex was produced ((Figure 5) B-7).
Figure 5: Withaferin A (1) binding to AKR1C3. Panels B-1 to B-5 show unbinding simulation phases of withaferin A: B-1 is the farthest from binding site, B-5 is the closest to binding site, and B-6 is binding site phase. Hydrogen bonds are shown as green-dashed lines with indicated bond length and the residues involved in hydrophobic interactions are shown as red arcs. Panel B-7 depicts the AKR1C3 docking in cartoon representation, where withaferin A (1) is shown as green sticks. The interacting residues are labeled and shown as surface of protein in different colors.
Unbinding simulation of AKR1C3/withalongolide A complex
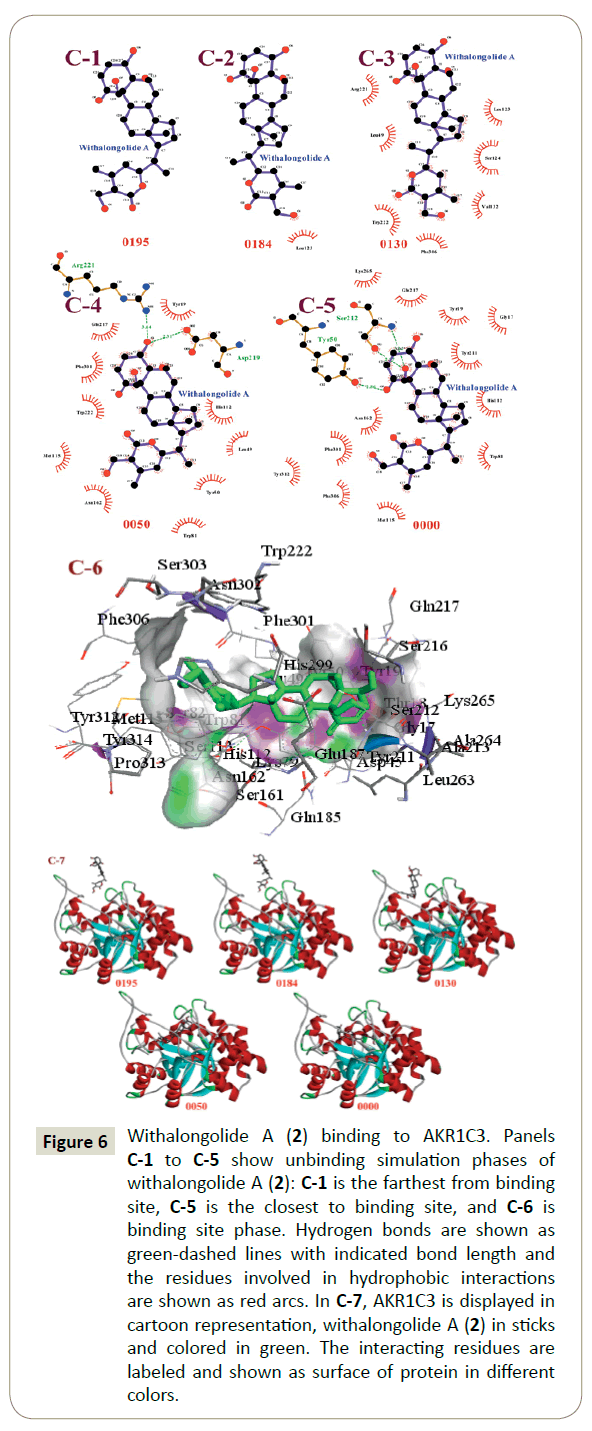
The unbinding simulation phases of withalongolide A (2) with increasing number of molecular interactions, while progressively approaching towards the binding site of AKR1C3 are shown in Figure 6 (C-1 to C-5). The docking phase displays the molecular interactions of AKR1C3 with withalongolide A (2), bound on the exterior region of AKR1C3 cavity by hydrophobic interactions. In addition, withalongolide A (2) was involved in interactions with the Leu 123 residue. The Leu 123 amino acid was common in all phases of unbinding simulations of withalongolide A (2), which emphasized the importance of Leu 123 in stabilizing AKR1C3- withalongolide A complex. A graphical representation of AKR1C3 in complex with withalongolide A (2) was generated ((Figure 6) C-6) and a cartoon representation of the dynamic simulations that display the movement and binding of withalongolide A with AKR1C3 to form the AKR1C3-withalongolide A complex was produced ((Figure 6) C-7).
Figure 6: Withalongolide A (2) binding to AKR1C3. Panels C-1 to C-5 show unbinding simulation phases of withalongolide A (2): C-1 is the farthest from binding site, C-5 is the closest to binding site, and C-6 is binding site phase. Hydrogen bonds are shown as green-dashed lines with indicated bond length and the residues involved in hydrophobic interactions are shown as red arcs. In C-7, AKR1C3 is displayed in cartoon representation, withalongolide A (2) in sticks and colored in green. The interacting residues are labeled and shown as surface of protein in different colors.
Unbinding simulation of AKR1C3/withalongolide B complex
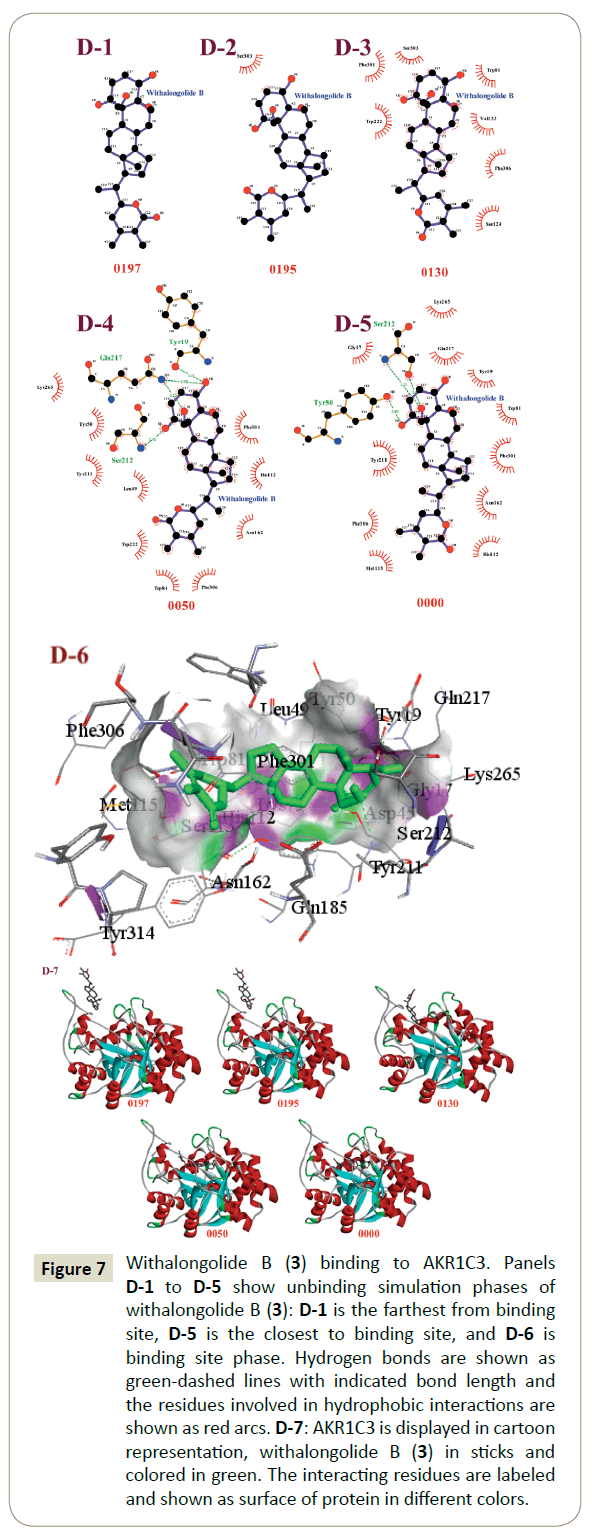
The unbinding simulation phases of withalongolide B (3) with increasing number of molecular interactions, while progressively approaching towards the binding site of AKR1C3 were conducted ((Figure 7) D-1 to D-5). The docking phase displays the molecular interactions of AKR1C3 with withalongolide B (3), bound on the exterior region of the AKR1C3 cavity by hydrophobic interactions. In addition, withalongolide B (3) was involved in interaction through the Asp 304 residue on AKR1C3. This amino acid was common in all phases of unbinding simulations of withalongolide B (3), which emphasized the importance of Asp 304 in stabilizing the AKR1C3/withalongolide B complex. A graphical representation of AKR1C3 in complex with withalongolide B (3) was generated ((Figure 7) D-6) and a cartoon representation of the dynamic simulations that display the movement and binding of withalongolide B with AKR1C3 to form the AKR1C3- withalongolide B complex was produced ((Figure 7) D-7).
Figure 7: Withalongolide B (3) binding to AKR1C3. Panels D-1 to D-5 show unbinding simulation phases of withalongolide B (3): D-1 is the farthest from binding site, D-5 is the closest to binding site, and D-6 is binding site phase. Hydrogen bonds are shown as green-dashed lines with indicated bond length and the residues involved in hydrophobic interactions are shown as red arcs. D-7: AKR1C3 is displayed in cartoon representation, withalongolide B (3) in sticks and colored in green. The interacting residues are labeled and shown as surface of protein in different colors.
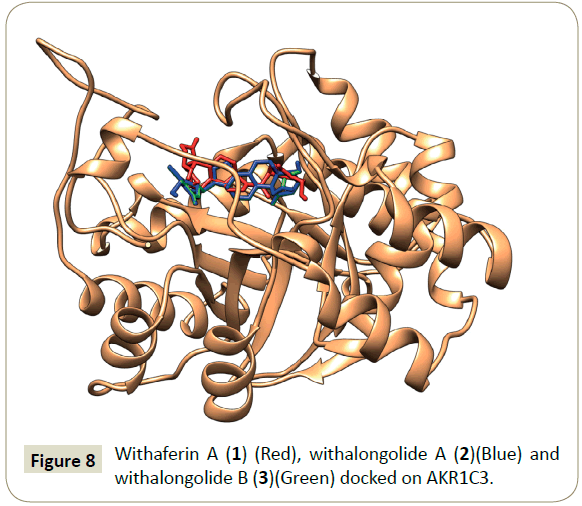
In summary, withalongolide B (3) docked to AKR1C3 with a better binding energy (-13.1 K cal), compared to other docked ligands: withalongolide A (2) (-12.5 K cal) and withaferin A (1) (-11.2 K cal). In addition, the MoMA server assisted unbinding simulation studies of AKR1C3/withaferin A, AKR1C3/withalongolide A and AKR1C3/withalongolide B complexes suggested that withalongolide B (3) took a longer time to unbind from AKR1C3 complex, compared to withalongolide A (2) and withaferin A (1). Based on this information, withalongolide B (3) was selected for further investigations in order to evaluate its effectiveness as a competent inhibitor of AKR1C3. A graphical representation of docked withaferin A (1), withalongolide A (2) and withalongolide B (3) in complex with AKR1C3 was generated (Figure 8).
Conclusion
The steroidogenic enzyme, AKR1C3, plays a critical role in prostate tumor androgen biosynthesis. In spite of the discovery of novel anticancer agents, drug resistance still proves to be a major problem in the successful treatment of solid tumors, which emphasizes the need for new and effective therapeutic targets. AKR1C3 is over expressed in various tumors in humans. Deregulated expression of AKR1C3 in multiple types of cancer contributes to the development of human cancer and chemotherapeutic resistance, which was primary cause of treatment failure. Flufenamic acid and indomethacin inhibited AKR1C3-dependent processes in human cell lines and murine xenografts of prostate cancer. However, the therapeutic efficacy of both flufenamic acid and indomethacin was restricted as a result of the undesired side effects that are associated with chronic cyclooxygenase enzymes. [32,33]. Our present in silico docking study suggested that withaferin A (1) withalongolide A (2) and withalongolide B (3) displayed strong interactions with AKR1C3 through hydrogen bonds, van der Waals and alkyl forces with binding energies (Kcal) of -11.2, -12.5 and -13.1 kcal/ mol, respectively. Additionally, unbinding simulation studies on the withanolides (1-3) in complex with AKR1C3 revealed that withalongolide B (3) took a longer time to unbind from AKR1C3, compared to withaferin A (1) or withalongolide A (2). These data also supported our in silico observations for withalongolide B (3). The current docking investigations strongly supports future research to evaluate the potential of withaferin A (1), withalongolide A (2) and withalongolide B (3) as AKR1C3 inhibitors in the treatment of drug resistance cancers. Combined, these results suggested among the withanolides investigated that withalongolide B (3) might function as the better AKR1C3 inhibitor and should be explored further as a potential solution to the abiraterone-resistance observed in prostate cancer.
References
- Khanna M, Qin KN, Wang RW, Cheng KC (1995) Substrate specificity gene structure and tissue-specific distribution of multiple human 3 alpha-hydroxystreoid dehydrogenases. J Biol Chem 270: 20162-20168.
- Matsuura K, Shiraishi H, Hara A, Sato K, Deyashiki Y, et al. (1998) Identification of a principle mRNA species for human 3alpha-hydroxysteroid dehydrogenase isoform (AKR1C3) that exhibits high prostaglandin D2 11-ketoreductse activity. Biochem 124: 940-946.
- Liu C, Armstrong CM, Lou W, Lombard A, Evans CP, et al. (2017) Inhibition of AKR1C3 activation overcomes resistance to abiraterone in advanced prostate cancer. Mol Cancer Ther 16: 35-44.
- Khanim F, Davies N, Veliça P, Hayden R, Ride J, et al. (2014) Selective AKR1C3 inhibitors do not recapitulate the anti-leukaemic activities of the pan-AKR1C inhibitor medroxyprogesterone acetate. Br J Cancer 110: 1506-1516.
- Samadi AK, Bazzill J, Zhang X, Gallagher R, Zhang H, et al. (2012) Novel withanolides target medullary thyroid cancer through inhibition of both RET phosphorylation and the mammalian target of rapamycin pathway. Surgery 152: 1238-1247.
- Lee I, Choi BY (2016) Witahferin-A-A natural anticancer agent with pleitropic mechanisms of action. Int J Mol Sci 17: 290.
- Dhami J, Chang E, Gambhir S (2016) Withaferin A and its potential role in glioblastoma (GBM). J Neuroncol 131: 201-211.
- Grogan PT, Sleder KD, Samadi AK, Zhang H, Timmermann BN, et al. (2013) Cytotoxicity of withaferin A in glioblastomas involves induction of an oxidative stress-mediated heat shock response while altering Akt/mTOR and MAPK signaling pathways. Invest New Drugs 31: 545-557.
- Zhang H, Samadi AK, Gallagher RJ, Araya JJ, Tong X, et al. (2011) Cytotoxic withanolide constituents of Physalis longifolia. J Nat Prod 74: 2532-2544.
- Gu M, Yu Y, Kamal GM, Gunaherath B, Gunatilaka AAL, et al. (2014) Structure-activity relationship (SAR) of withanolides to inhibit Hsp90 for its activity in pancreatic cancer cells. Invest New Drugs 32: 68-74.
- Hahm E, J Lee J, Huang Y, Singh SV (2011) Withaferin A suppresses estrogen receptor-α expression in human breast cancer cells. Mol Carcinog 50: 614-624.
- Grover A, Priyandoko D, Gao R, Shandilya A, Widodo N, et al. (2012) Withanone binds to mortlin and abrogates mortalin-p53 complex : computational and experimental evidence. Int J Biochem Vell Biol 44: 496-504.
- Vaishnavi1 K, Saxena N, Shah N, Singh R, Uthayakumar1 KMM, et al. (2012) Differential activities of the two closely related withanolides, withaferin A and withanone: Bioinformatics and Experimental Evidences. PLoS One 7: e44419.
- Yadav DK, Kumar S, Saloni S, Singh H, Kim M, et al. (2017) Molecular docking, QSAR and ADMET studies of withanolide analogues against breast cancer. Drug Dis Devel Ther 11: 1859-1870.
- Remya C, Dileep KV, Variayr EJ, Sadasivan C (2016) An in silico guided identification of nACHR agonists from Withanifera somnifera. Frontiers Life Sci 9: 201-213.
- Puspaningtyas AR (2014) Docking studies of Physalis peruviana ethanol extract using molegro virtual docker on insulin tyrosine kinase receptor as antidiabetic agent. Int Curr Pharm J 3: 265-269.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46: 3-26.
- Chothia C, Lesk AM (1986) The relation between the divergence of sequence and structure in proteins. EMBO J 5: 823-826.
- Kaczanowski S, Zielenkiewicz P (2010) Why similar protein sequences encode similar three-dimentional structures? Theor Chem Acc 125: 643-650.
- Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, et al. (2008) Protein structure homology modelling using SWISS-MODEL workspace. Nature Protocols 4: 1-13.
- Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31: 3381-3385.
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, et al. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42: 195-201.
- Read RJ, Adams PD, Arendall III WB, Brunger AT, Emsley P, et al. (2011) A New generation of crystallographic validation tools for the protein data bank. Structure 19: 1395-1412.
- Savarino A (2007) In Silico docking of HIV-1 integrase inhibitors reveals a novel drug type acting on an enzyme/DNA reaction intermediate. Retrovirol 4: 1-21.
- Nousheen L, Akkiraju PC, Enaganti S (2014) Molecular docking mutational studies on human surfactant protein-D. World J Pharmaceut Res 3: 1140-1148.
- Jin H, Zhou Z, Wang D, Guan S, Han W (2015) Molecular dynamics simulations of acylpeptide hydrolase bound to chlopyrifosmethyl oxon and dichlorvos. Inter J Mol Sci 16: 6217-6234.
- Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, et al. (2012) Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J Cheminform 4: 17.
- Dallakyan S, Oleson A (2015) Small-molecule library screening by docking with PyRx Methods. Mol Biol 1263: 243-250.
- Devaurs D, Bouard L, Vaisset M, Zanon C, Al-Bluwi I, et al. (2013) MoMA-LigPath: a web server to simulate protein-ligand unbinding. Nucleic Acids Res 41: W297-W302.
- Cortes J, Le DT, Iehl R, Simeon T (2010) Simulating ligand-induced conformational changes in proteins using a mechanical disassembly method. Physical Chem Chem Phys 12: 8268-8276.
- Penning TM (2015) Mechanisms of drug resistance that target the androgen axis in castration resistant prostate cancer (CRPC). J Steroid Biochem Mol Biol 153: 105-113.
- Adeniji AO, Chen M, Penning TM (2013) AKR1C3 as a target in castrate resistant prostate cancer. J Steroid Biochem Mol Biol 137: 136-149.
- Zhao J, Xiang Y, Xiao C, Guo P, Wang D, et al. (2014) AKR1C3 overexpression mediates methaotrexate resistance ib choriocarcinoma cells. Int J Med Sci 11: 1089-1097.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences