An In Silico Approach for the Evaluation of Nutraceutical Properties of Pearl Millet as a Brain Food
Rathore S, Verma S, Singh K and Pandey AK
DOI10.21767/2469-6692.100018
Rathore S1, Verma S2, Singh K3 and Pandey AK4*
1Department of Food Technology, Hartcourt Butler Technical University, Kanpur, Uttar Pradesh, India
2Department of Biotechnology, Institute of Engineering and Technology, APJ Abdul Kalam Technical University, Lucknow, Uttar Pradesh, India
3Department of Food Technology, School of Chemical Technology, Hartcourt Butler Technical University, Kanpur, Uttar Pradesh, India
4Department of Biotechnology, Institute of Engineering and Technology, Bundelkhand University, Jhansi, Uttar Pradesh, India
- *Corresponding Author:
- Pandey AK
Assistant Professor
Department of Biotechnology Engineering
Institute of Engineering and Technology
Bundelkhand University Jhansi
Uttar Pradesh, India
Tel: +91 9030961817
E-mail: vnsanand_9@rediffmail.com
Received Date: June 07, 2017; Accepted Date: June 29, 2017; Published Date: July 06, 2017
Citation: Rathore S, Verma S, Singh K, Pandey AK (2017) An In Silico Approach for the Evaluation of Nutraceutical Properties of Pearl Millet as a Brain Food. J In Silico In Vitro Pharmacol. 3:18. doi: 10.21767/2469-6692.10018
Abstract
Introduction: Pearl millet’s high flavonoid content has incredible anti-inflammatory and anti-oxidant properties to reduce oxidative stress and inflammation, common pathologies of Neurodegenerative Diseases (NDs). Researches have proved that MMPs, ASIC1a, AQP-4 and PKC plays major role in most NDs and their structural inhibition can provide an effective treatment.
Methods and Findings: Molecular docking study of pearl millet’s constituents (Ellagic acid, gallic acid, tryptophan, linolenic acid, quercetin, pelargonidin, vitexin, glycosylorientin and glycosylvitexxin) with MMP9, MMP2, Aquaporin4, ASIC1a, and PKC was performed using Autodock software. Quercetin showed significant inhibition of MMP9 having binding energy of -10.41 kcal/mol with 2 H-bonds, MMP2: -9.32 kcal/mol with 4 H-bonds, ASIC1a: -6.4 kcal/mol with 4H-bonds, Aquaporin 4: -7.57 kcal/mol with 3-H bonds and PKC: -6.38 kcal/mol with 1-H bonds. Inhibitory binding of MMP9, MMP2, Aquaporin4, ASIC1a and PKC with; pelargonidin formed 2, 4, 3, 1 and 2 H-bonds with binding energies of -9.3, -7.16, -10.34, -5.95 and -6.84 kcal/mol respectively; vitexin formed 4, 3, 4, 3 and 5 H-bonds with binding energies of -7.71, -8.63, -6.01, -7.07 and -4.97 kcal/mol respectively; glycosylorientin: 7, 6, 3, 2 and 3 H-bonds with binding energies of -6.15, -4.15, -3.51, -5.13 and -4.62 kcal/mol respectively and glycosylvitexin: 4, 3, 2, 3 and 1 H-bonds having binding energies of -5.51, -6.29, -5.07, -4.63 and -6.11 kcal/mol respectively.
Conclusion: Pearl millet can efficiently treat NDs through regular diet and extensive research is required to exploit its potential to develop specifically targeted drugs for specific NDs.
Keywords
Pearl millet; Molecular docking; Neurodegenerative diseases; Anti-oxidants
Abbreviations
MMP: Matrix Metallo Proteinase; ASIC1a: Acid Sensing Ion Channel 1a; AQP4: Aquaporin 4; PKC: Protein Kinase C
Introduction
Treatment through regular diet can increase the efficiency and ease of NDs treatment. Nutraceuticals are the components of food taken in regular diet which not only provide nutrition but also pose medicinal effects proving their cumulative beneficiary effect for health [1]. Polyphenols including flavonoids are potent nutraceutical, which have high anti-oxidant properties and can help to cure most diseases associated with oxidative stress [2]. Coarse grains mainly grown in Asian and African tropical semi-arid regions include pearl millet, sorghum, oats, barley and maize as well as other millets has high nutritional value. They can provide good supply of energy, minerals, vitamins, fibre content and most importantly have high flavonoid content providing them anti-oxidant property [3].
Pearl millet (Pennisetum glaucum) or spiked millet of family Gramineae, commonly grown in tropics and subtropics of Asia and Africa has a high nutritive value than that of wheat, rice, sorghum or maize. It has enough amounts of zinc, iron and high flavonoid content enhancing anti-oxidant and anti-inflammatory properties [4]. In turn, pearl millet has glycaemic index of 55 which is less as compared to other millets, provides high energy of 361 kcal per 100 g, the higher fibre content of 1.2 g per 100 g and carbohydrate content of 67.5 g per 100 g which is comparatively less than rice, sorghum and wheat but greater than maize. Along with this, low starch which is gluten free, circa 8 to 15 times activity of α-amylase in comparison of wheat, and high oil and protein content ranging from 8 to 19% on an average 11.6% with low levels of tryptophan, lysine, threonine and sulphur containing amino acid, enhances its nutraceutical properties [5,6]. It also has high fat content which is 5 mg per 100 g comprising of 75% fatty acids which are unsaturated. Good fatty acids like Omega 3 and linolenic acids accounting for 4% of fatty acids boost its nutritive quality [7]. All these qualities of pearl millet make it a good food for treating gastrointestinal disorders, cardiovascular problems, diabetes as well as cancer [5,8]. The strong nutraceutical components of pearl millet contributing for it anti-oxidant and anti-inflammatory properties are pelargonidin, quercetin, tannin (Ester of gallic acid and ellagic acid) and phytic acid [8]. Various other phenolic compounds are vitexin, glucosylvitexin and glucosylorientin, all these together contribute to combat oxidative stress which is common to the pathology of most of the neurodegenerative diseases [2].
Neurodegenerative Disorders (NDs), due to lack of knowledge about pathology and out of reach drug targets are increasing rapidly in population around the world. Due to difficulty in passing the blood brain barrier most of the treatments till date are facing a great challenge [9]. The increased incidence of NDs in society is making an urgent demand for effective treatments. Most of the NDs are having oxidative stress as a common pathological feature. NDs like cerebral ischemia, Alzheimer’s disease, Parkinson’s disease, multiple sclerosis, depression, etc. has been reported to have oxidative stress as a major cause for neuronal damage [1]. Oxygen is utmost important for life, but its consumption leads to generation of free radicals. Free radicals are produced in the body as regular by products from various mechanisms. These free radicals comprise potential to cause damage to the environment by reacting with the healthy cellular organisation and break them down [10]. But the body’s antioxidant level prevents them from causing such damage. Under diseased condition, due to mutations or injuries or any other possible reason; the degeneration of cells and accumulation of oxidant in cellular environment cause the disturbance in the antioxidant defence mechanism, due to which cells losses the capability of their normal functioning and leads to apoptosis [10]. As it is difficult to balance this antioxidant amounts from external sources in case of NDs due to lack of known targets and inability of compounds to cross defensive mechanism like blood brain barrier, it is better to supply them through dietary sources [11]. So, in this study, we hypothesize that pearl millet being a good source of antioxidants can be used for the reason, as it has a large variety of components like pelargonidin, quercetin, tannin, glycosylvitexin, vitexin, glycosylorientin and phytic acid [5]. The presence of anti-inflammatory components of pearl millets such as Omega-3 and linolenic acids enhances its therapeutic potential against NDs like Alzheimer’s and multiple sclerosis respectively [8].
To strengthen our hypothesis we performed in silico molecular docking to detect for direct structural inhibition provided by these compounds to major targets of NDs. We targeted different molecular targets i.e., proteins that involved in various NDs with the above-mentioned components of pearl millet as inhibitors. It has been earlier reported by Bhattacharya et al. [12] that cerebral ischemia involves interruption in the blood supply to the brain leading to neuronal death which causes acidosis and the activation of Matrix Metalloproteinase (MMP). Acidosis also leads to the activation of acid sensing ion channels 1a (ASIC 1a) which further leads to the activation of μ-calpain having cell suicidal capabilities by fragmentation of cytoskeleton protein “spectrin” [12]. Brain edema is a major pathological feature of ischemic injury and Aquaporin 4 is the water transporting channel which plays a major role in enhancing edema and hence the severity of disease [13]. In Alzheimer’s disease, due to impairment of synaptic plasticity, memory loss is considered a major pathological feature and protein kinase C (PKC) has prominent role in maintaining synaptic plasticity along with memory persistence function. PKC also regulate the processing of amyloid precursor protein, a major hallmark of Alzheimer’s. Also, there is a decrease in the level of MMP2 and increase in level of MMP9 found in Alzheimer’s brain which is capable of regulating amyloid beta accumulations [14,15]. Aquaporin 4 also plays a great role in Alzheimer’s pathology as it affects synaptic plasticity, memory impairment and also involved in amyloid beta clearance. So, the dysregulation of the above mentioned can cause the degenerative effects [16]. In the case of Parkinson’s disease also, these molecular targets contain treatment potency e.g. PKC is also found to play role in Parkinson’s pathology. Another one is multiple sclerosis in which levels of MMP9 are elevated and becomes the cause of relapsing condition in short duration [17]. ASIC1a is also known to express increasingly and are a potent target for multiple sclerosis as they are responsible for the accumulation of intracellular ions of sodium and calcium and causes injurious symptoms of the disease [18]. So, we considered MMP9, MMP2, Aquaporin 4, ASIC1a and PKC as major targets for our in silico study on NDs treatment.
Materials and Methods
The method implemented in our analysis was molecular docking by the use of Autodock software. Targets used were MMP9, MMP2, Aquaporin-4, ASIC1a and Protein Kinase C, their crystal structures were taken from RCSB protein data bank (PDB id. 1L6J, 1QIB, 2D57, 3HGC and 1XJD respectively). The structures of ligands i.e., pelargonidin, quercetin, ellagic acid, gallic acid, vitexin, glycosylorientin, glycosylvitexin and linolenic acid were made by using ACD Lab and the PDB files of all ligands were made by using online smile translator. By using Autodock tools, firstly the grid box was generated that was enough to capture the entire protein’s catalytic site and also can accommodate all above mentioned ligands to move freely during docking. The total Kolhman’s charges added were -207.662, -83.024, -86.244, -247.389 and -143.943 to different PDB i.e., 1L6J, 1Q1B, 2D57, 3HGC and 1XJD respectively. Lamarckian genetic search algorithm was employed and thirty search attempts (GA run parameter) were performed, with a population size of 150. The lamarkian GA parameters used in the study were numbers of runs 2500000, the number of generation 27000, the rate of gene mutation 0.02 and the cross over rate is 0.8. Blind docking was done with the grid size of 126, 126, and 126 along the three axes X, Y, and Z respectively, with 0.375 Å spacing. The grid centre was set to 50.728, 9.883, and 6.739 Å. Other docking parameters were set to the software’s default values. After docking, the ligands were ranked according to their docked or binding energy as used in the Autodock software. Close by views of the interactions of binding of all the targets with the ligands were analyzed through Python Molecular viewer. The primary screening on the basis of the molecular properties and bioactivity score of the ligands was done by using Molinspiration tool implementing smile notation as input.
Results
In our docking study we found many constituents of pearl millet that showed effective docking with the proteins involved in various neurodegenerative diseases. The constituents that were considered as inhibitory ligands were selected on the basis of molecular properties and bioactivity scores, which gave an idea about the ligands effectiveness against NDs. The parameters that are essential for neurological drugs are logP value which is required to be less than 7, greater TPSA or surface area and high affinity for GPCR ligands. According to all these parameters we found pelargonidin, quercetin, ellagic acid and gallic acid (which on esterification forms tannins), glycosylvitexin, glycosylorientin, vitexin, tryptophan and linolenic acid to be considered for docking studies (Table 1). The potent targets that we considered were MMP9 (PDB 1L6J), MMP2 (PDB 1Q1B), Aquaporin 4 (PDB 2D57), ASIC1a (PDB 3HGC) and Protein kinase C (PDB 1XJD) as they play major role in NDs like cerebral ischemia, Alzheimer’s, Parkinson’s, multiple sclerosis and depression according to past researches.
| S.No. | Name | Structure | Mol.Wt. (g/mol) |
IUPAC | Smile Notation | Target | Molecular properties /Bioactivity Score | Amount (mg/100g) |
|
|---|---|---|---|---|---|---|---|---|---|
| 1 | Tannic acid (Ester of gallic acid and ellagic acid) |
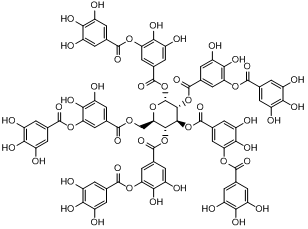 C76H52O46 |
1701.19 | 2,3-dihydroxy-5-({[(2R,3R,4S,5R,6R)-3,4,5,6-tetrakis({3,4-dihydroxy-5-[(3,4,5-trihydroxyphenyl)carbonyloxy]phenyl}carbonyloxy)oxan-2-yl]methoxy}carbonyl)phenyl 3,4,5-trihydroxybenzoate | Oc1cc(cc(O)c1O)C(=O)Oc1cc(cc(O)c1O)C(=O)OC[C@H]1O[C@H](OC(=O)c2cc(O)c(O)c (OC(=O)c3cc(O)c(O)c(O)c3)c2)[C@](OC(=O)c2cc(O)c(O)c(OC(=O)c3cc(O)c(O)c(O)c3)c2)[C@@H](OC(=O)c2cc(O)c(O)c(OC(=O)c3cc(O)c(O)c(O)c3)c2)[C@@H]1OC(=O)c1cc(O)c(O)c( OC(=O)c2cc(O)c(O)c(O)c2)c1 |
Cancer, Alzheimer’s Parkinson’s, multiple sclerosis | Mol. Properties | 199{Azhari A.M. NOUR, 2014 #63} | |
| miLogP | 7.06 | ||||||||
| TPSA | 777.98 | ||||||||
| Natoms | 122 | ||||||||
| MW | 1701.21 | ||||||||
| nON | 46 | ||||||||
| nOHNH | 25 | ||||||||
| Nviolations | 4 | ||||||||
| Nrotb | 31 | ||||||||
| Volume | 1305.93 | ||||||||
| Bio. Score | |||||||||
| GPCR ligand | -6.10 | ||||||||
| Ion channel modulator | -6.24 | ||||||||
| Kinase inhibitor | -6.31 | ||||||||
| Nuclear receptor ligand | -6.35 | ||||||||
| Protease inhibitor | -5.99 | ||||||||
| Enzyme inhibitor | -6.07 | ||||||||
| 1a | Gallic acid | 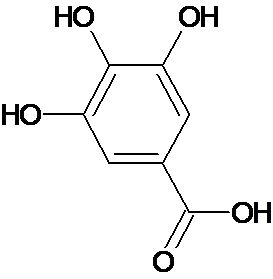 C7H6O5 |
170.12 g | 3,4,5-Trihydroxybenzoic acid | O=C(O)c1cc(O)c(O)c(O)c1 | Neurodegenerative diseases, Parkinson’s mensural problems, cardiovascular problems, cancer, renal diseases, microbial infection and depression | Mol. Properties | ||
| miLogP | 0.59 | ||||||||
| TPSA | 97.98 | ||||||||
| Natoms | 12 | ||||||||
| MW | 170.12 | ||||||||
| nON | 5 | ||||||||
| nOHNH | 4 | ||||||||
| Nviolations | 0 | ||||||||
| nrotb | 1 | ||||||||
| volume | 135.10 | ||||||||
| Bio. Score | |||||||||
| GPCR ligand | -0.77 | ||||||||
| Ion channel modulator | -0.26 | ||||||||
| Kinase inhibitor | -0.88 | ||||||||
| Nuclear receptor ligand | -0.52 | ||||||||
| Protease inhibitor | -0.94 | ||||||||
| Enzyme inhibitor | -0.17 | ||||||||
| 1b | Ellagic Acid | 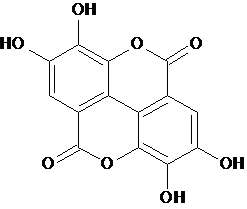 C14H6O8 |
302.197 | 2,3,7,8-Tetrahydroxy-chromeno[5,4,3-cde]chromene-5,10-dione | O=C1Oc3c2c4c1cc(O)c(O)c4OC(=O)c2cc(O)c3O | Mol. Properties | |||
| miLogP | 0.94 | ||||||||
| TPSA | 141.33 | ||||||||
| Natoms | 22 | ||||||||
| MW | 302.19 | ||||||||
| nON | 8 | ||||||||
| nOHNH | 4 | ||||||||
| Nviolations | 0 | ||||||||
| nrotb | 0 | ||||||||
| volume | 221.78 | ||||||||
| Bio. Score | |||||||||
| GPCR ligand | -0.29 | ||||||||
| Ion channel modulator | -0.27 | ||||||||
| Kinase inhibitor | -0.01 | ||||||||
| Nuclear receptor ligand | 0.11 | ||||||||
| Protease inhibitor | -0.18 | ||||||||
| Enzyme inhibitor | 0.17 | ||||||||
| 2 | Pelargonidin | 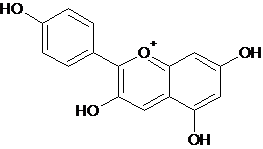 C15H11O5+ |
271.24 |
2-(4-Hydroxyphenyl)chromenylium-3,5,7-triol | C1=CC(=CC=C1C2=C(C=C3C(=CC(=CC3=[O+]2)O)O)O)O | Allergic disease, ophthalmic diseases Heart diseases, diabetes milletus, urinary infection |
Mol. Properties | 3.310 | |
| miLogP | -0.26 | ||||||||
| TPSA | 92.08 | ||||||||
| Natoms | 20 | ||||||||
| MW | 271.25 | ||||||||
| nON | 5 | ||||||||
| nOHNH | 4 | ||||||||
| Nviolations | 0 | ||||||||
| nrotb | 1 | ||||||||
| volume | 226.79 | ||||||||
| Bio. Score | |||||||||
| GPCR ligand | -0.18 | ||||||||
| Ion channel modulator | -0.11 | ||||||||
| Kinase inhibitor | -0.07 | ||||||||
| Nuclear receptor ligand | 0.03 | ||||||||
| Protease inhibitor | -0.33 | ||||||||
| Enzyme inhibitor | -0.02 | ||||||||
| 3 | Quercetin | 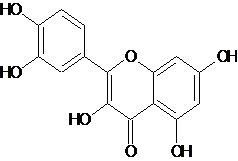 C15H10O7 |
302.236 | 2-(3,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-one | O=C1c3c(O/C(=C1/O)c2ccc(O)c(O)c2)cc(O)cc3O | Alzhiemers, Parkinsons, cerebralischemia , multiple sclerosis, heart diseases, common cold, heart diseases, cancer and skin and tissue diseases, anti thrombiotic and antiinfllamatory , obesity related disorders | Mol. Properties | 10.210 | |
| miLogP | 1.68 | ||||||||
| TPSA | 131.35 | ||||||||
| Natoms | 22 | ||||||||
| MW | 302.24 | ||||||||
| nON | 7 | ||||||||
| nOHNH | 5 | ||||||||
| Nviolations | 0 | ||||||||
| nrotb | 1 | ||||||||
| volume | 240.08 | ||||||||
| Bio. Score | |||||||||
| GPCR ligand | -0.06 | ||||||||
| Ion channel modulator | -0.19 | ||||||||
| Kinase inhibitor | 0.28 | ||||||||
| Nuclear receptor ligand | 0.36 | ||||||||
| Protease inhibitor | -0.25 | ||||||||
| Enzyme inhibitor | 0.28 | ||||||||
| 4 | Phytic acid | 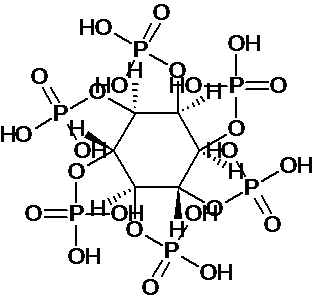 C6H18O24P6 |
660.03 |
(1R,2R,3S,4S,5R,6S)-cyclohexane-1,2,3,4,5,6-hexayl hexakis[dihydrogen (phosphate)] | [C@@H]1([C@@H]([C@@H]([C@@H]([C@H]([C@@H]1OP(=O)(O)O)OP(=O)(O)O)OP(=O)(O)O)OP(=O)(O)O)OP(=O)(O)O)OP(=O)(O)O | Alzhiemers, parkinsons, multiple sclerosis, colon cancer and serum cholesterol | Mol. Properties | 203.029 | |
| miLogP | -5.55 | ||||||||
| TPSA | 400.57 | ||||||||
| Natoms | 36 | ||||||||
| MW | 660.03 | ||||||||
| nON | 24 | ||||||||
| nOHNH | 12 | ||||||||
| Nviolations | 3 | ||||||||
| nrotb | 12 | ||||||||
| volume | 422.96 | ||||||||
| Bio. Score | |||||||||
| GPCR ligand | 0.38 | ||||||||
| Ion channel modulator | 0.44 | ||||||||
| Kinase inhibitor | 0.37 | ||||||||
| Nuclear receptor ligand | 0.24 | ||||||||
| Protease inhibitor | 0.31 | ||||||||
| Enzyme inhibitor | 0.48 | ||||||||
| 5 | Tryptophan | 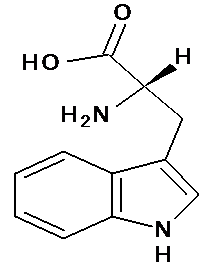 C11H12N2O2 |
204.23 | Tryptophan or (2S)-2-amino-3-(1H-indol-3-yl) propanoic acid | c1ccc2c(c1)c(c[nH]2)C[C@@H](C(=O)O)N | Increases serotonin levels anti-depressent Have anti -oxidant property | Mol. Properties | tryptophan content in prolamins protein(.0024mg/100g)and non-prolamin proteins (.0012–.0014mg/100g)11 | |
| miLogP | -1.08 | ||||||||
| TPSA | 79.11 | ||||||||
| Natoms | 15 | ||||||||
| MW | 204.23 | ||||||||
| nON | 4 | ||||||||
| nOHNH | 4 | ||||||||
| Nviolations | 0 | ||||||||
| nrotb | 3 | ||||||||
| volume | 184.94 | ||||||||
| Bio Score | |||||||||
| GPCR ligand | 0.33 | ||||||||
| Ion channel modulator | 0.54 | ||||||||
| Kinase inhibitor | -0.13 | ||||||||
| Nuclear receptor ligand | -0.22 | ||||||||
| Protease inhibitor | 0.15 | ||||||||
| Enzyme inhibitor | 0.44 | ||||||||
| 6 | Glucosyl-vitexin | 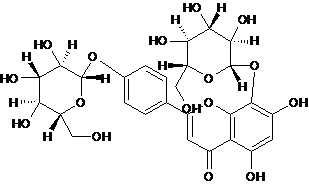 C27H30O16 |
610.517 | 5,7-dihydroxy-8-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl) oxan-2-yl]oxy-2-[4-[(2S,3R,4S,5S, 6R ) -3,4,5-trihydroxy-6-(hydroxyl methyl) oxan-2-yl] oxyphenyl ] chromen-4-one | C1=CC(=CC=C1C2=CC(=O)C3=C(O2)C(=C(C=C3O)O)O[C@H]4[C@@H]([C@H]([C@@H]([C@H](O4)CO)O)O)O)O[C@H]5[C@@H]([C@H]([C@@H]([C@H](O5)CO)O)O)O | Antioxidant | Mol. Properties | 17012 | |
| miLogP | -1.63 | ||||||||
| TPSA | 269.43 | ||||||||
| Natoms | 43 | ||||||||
| MW | 610.52 | ||||||||
| nON | 16 | ||||||||
| nOHNH | 10 | ||||||||
| Nviolations | 3 | ||||||||
| nrotb | 7 | ||||||||
| volume | 496.3 | ||||||||
| Bio. Score | |||||||||
| GPCR ligand | -0.05 | ||||||||
| Ion channel modulator | -0.42 | ||||||||
| Kinase inhibitor | -0.15 | ||||||||
| Nuclear receptor ligand | -0.11 | ||||||||
| Protease inhibitor | -0.04 | ||||||||
| Enzyme inhibitor | 0.08 | ||||||||
| 7 | Glucosyl-orientin | 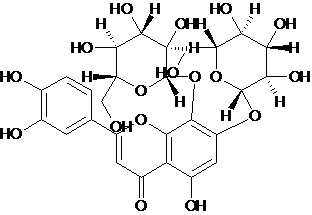 C27H30O17 |
626.516 | 2-(3,4-dihydroxyphenyl)-5-hydroxy-7,8-bis[[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy]chromen-4-one | C1=CC(=C(C=C1C2=CC(=O)C3=C(O2)C(=C(C=C3O)O[C@H]4[C@@H]([C@H]([C@@H]([C@H](O4)CO)O)O)O)O[C@H]5[C@@H]([C@H]([C@@H]([C@H](O5)CO)O)O)O)O)O | Antioxidant | Mol. properties | 64.7512 | |
| miLogP | -2.35 | ||||||||
| TPSA | 289.65 | ||||||||
| Natoms | 44 | ||||||||
| MW | 626.52 | ||||||||
| nON | 17 | ||||||||
| nOHNH | 11 | ||||||||
| Nviolations | 3 | ||||||||
| nrotb | 7 | ||||||||
| volume | 504.33 | ||||||||
| Bio. Score | |||||||||
| GPCR ligand | -0.11 | ||||||||
| Ion channel modulator | -0.54 | ||||||||
| Kinase inhibitor | -0.21 | ||||||||
| Nuclear receptor ligand | -0.21 | ||||||||
| Protease inhibitor | -0.08 | ||||||||
| Enzyme inhibitor | 0.01 | ||||||||
| 8 | Vitexin | 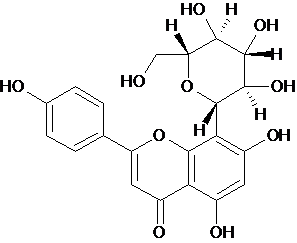 C21H20O10 |
432.377 | 5,7-dihydroxy-2-(4-hydroxyphenyl)-8-[(2S,3R,4R,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]chromen-4-one | C1=CC(=CC=C1C2=CC(=O)C3=C(O2)C(=C(C=C3O)O)[C@H]4[C@@H]([C@H]([C@@H]([C@H](O4)CO)O)O)O)O | Antioxidant | Mol. Properties | 23.5412 | |
| miLogP | 0.52 | ||||||||
| TPSA | 181.04 | ||||||||
| Natoms | 31 | ||||||||
| MW | 432.38 | ||||||||
| nON | 10 | ||||||||
| nOHNH | 7 | ||||||||
| Nviolations | 1 | ||||||||
| nrotb | 3 | ||||||||
| volume | 355.20 | ||||||||
| Bio. Score | |||||||||
| GPCR ligand | 0.13 | ||||||||
| Ion channel modulator | -0.14 | ||||||||
| Kinase inhibitor | 0.19 | ||||||||
| Nuclear receptor ligand | 0.23 | ||||||||
| Protease inhibitor | 0.03 | ||||||||
| Enzyme inhibitor | 0.46 | ||||||||
| 9 | Linolenic acid | 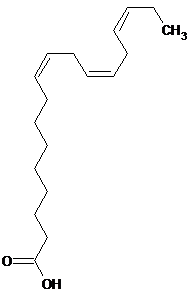 C18H30O2 |
278.44 | (9Z,12Z,15Z)-9,12,15-Octadecatrienoic acid | CC/C=C\C/C=C\C/C=C\CCCCCCCC(=O)O | Conditions of rheumatoid arthritis, cardiac arrhythmias, depression and reduce the risk of ischemic and thrombotic stroke. |
Mol. properties | 238413 | |
| miLogP | 5.84 | ||||||||
| TPSA | 37.30 | ||||||||
| Natoms | 20 | ||||||||
| MW | 278.44 | ||||||||
| nON | 2 | ||||||||
| nOHNH | 1 | ||||||||
| Nviolations | 1 | ||||||||
| nrotb | 13 | ||||||||
| volume | 306.47 | ||||||||
| Bio. Score | |||||||||
| GPCR ligand | 0.33 | ||||||||
| Ion channel modulator | 0.23 | ||||||||
| Kinase inhibitor | -0.19 | ||||||||
| Nuclear receptor ligand | 0.35 | ||||||||
| Protease inhibitor | 0.13 | ||||||||
| Enzyme inhibitor | 0.42 | ||||||||
Table 1: Constituent of pearl millet having antioxidant property. Different properties: (1) Antioxidant property; (2) Docking results.
Pelargonidin
The blind docking of pelargonodin with MMP9, MMP2, Aquaporin 4, ASIC1a, and PKC stabilized the target proteins with the binding energies of -10.34, -9.3, -7.16, -6.84 and -5.95 kcal/mol respectively which are potentially good to prove pelargonidin as an inhibitor and to evident its effectively for the neurodegenerative diseases in which these targets are involved (Table 2).
| MMP9 H bonds BE (kcal/mol) |
MMP2 H bonds BE (kcal/mol) |
Aquaporin 4 H bonds BE (kcal/mol) |
ASIC1a H bonds BE (kcal/mol) |
PKC H bonds BE (kcal/mol) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pelargonidin | 2 | -10.34 | 4 | -9.3 | 3 | -7.16 | 1 | -6.84 | 2 | -5.95 |
| Quercetin | 2 | -10.41 | 4 | -9.32 | 3 | -7.57 | 4 | -6.4 | 1 | -6.38 |
| Ellagic acid | 2 | -8.46 | 3 | -7.56 | 1 | -3.22 | 4 | -9.93 | 3 | -7.11 |
| Gallic acid | 2 | -4.27 | 3 | -4.44 | 2 | -4.19 | 3 | -4.46 | 2 | -4.1 |
| Vitexin | 4 | -7.71 | 3 | -8.63 | 4 | -6.01 | 3 | -7.07 | 5 | -4.97 |
| Glycosylvitexin | 4 | -5.51 | 3 | -6.29 | 2 | -5.07 | 3 | -4.63 | 1 | -6.11 |
| Glycosylorientin | 7 | -6.15 | 6 | -4.15 | 3 | -3.51 | 2 | -5.13 | 3 | -4.62 |
| Tryptophan | 2 | -7.86 | 3 | -8.13 | 2 | -5.84 | 3 | -6.62 | 1 | -5.58 |
| Linolenic acid | 0 | -5.42 | 0 | -6.24 | 2 | -4.51 | 2 | -5.64 | 1 | -4.3 |
Table 2: Docking results showing H-bonds and binding energy between different proteins involved in neurodegenerative disorders and the constituents of pearl millet.
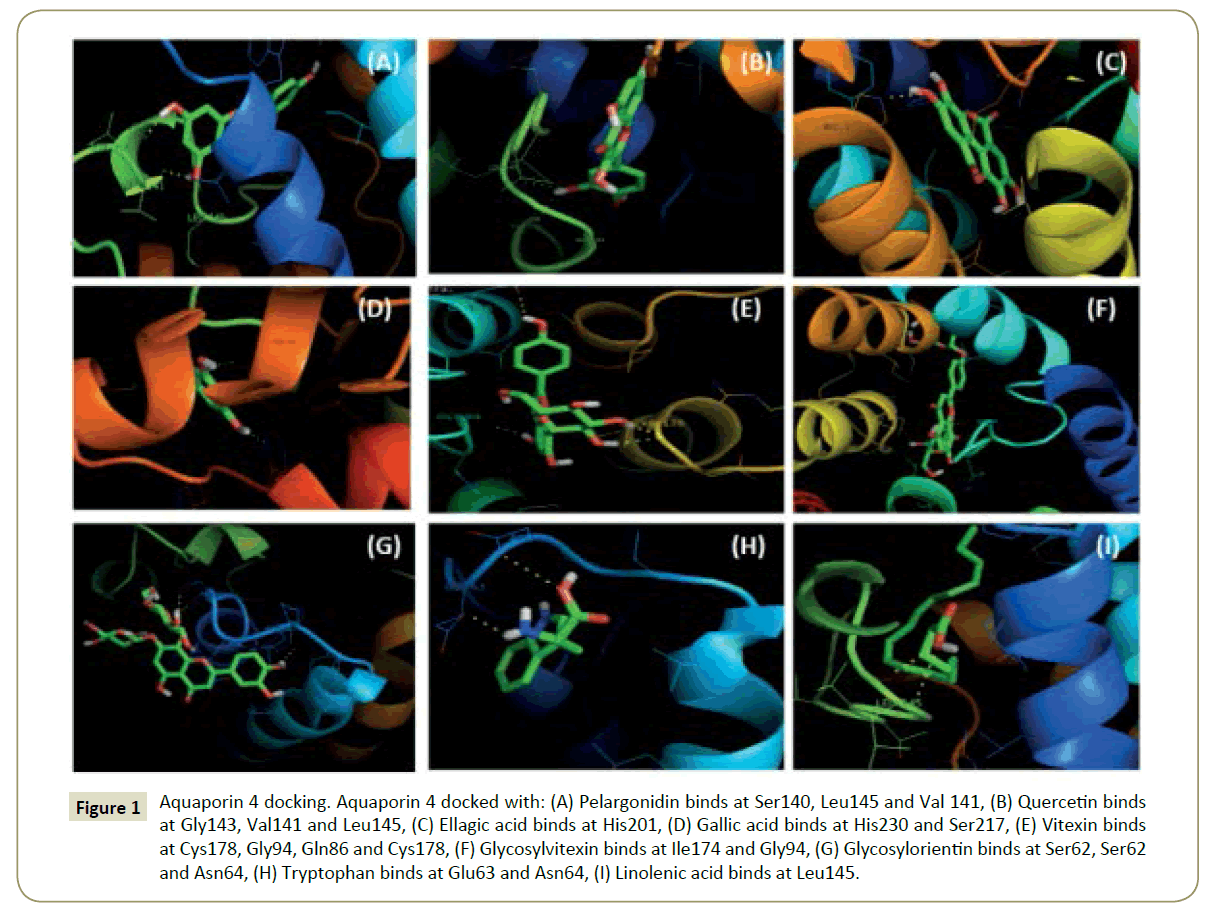
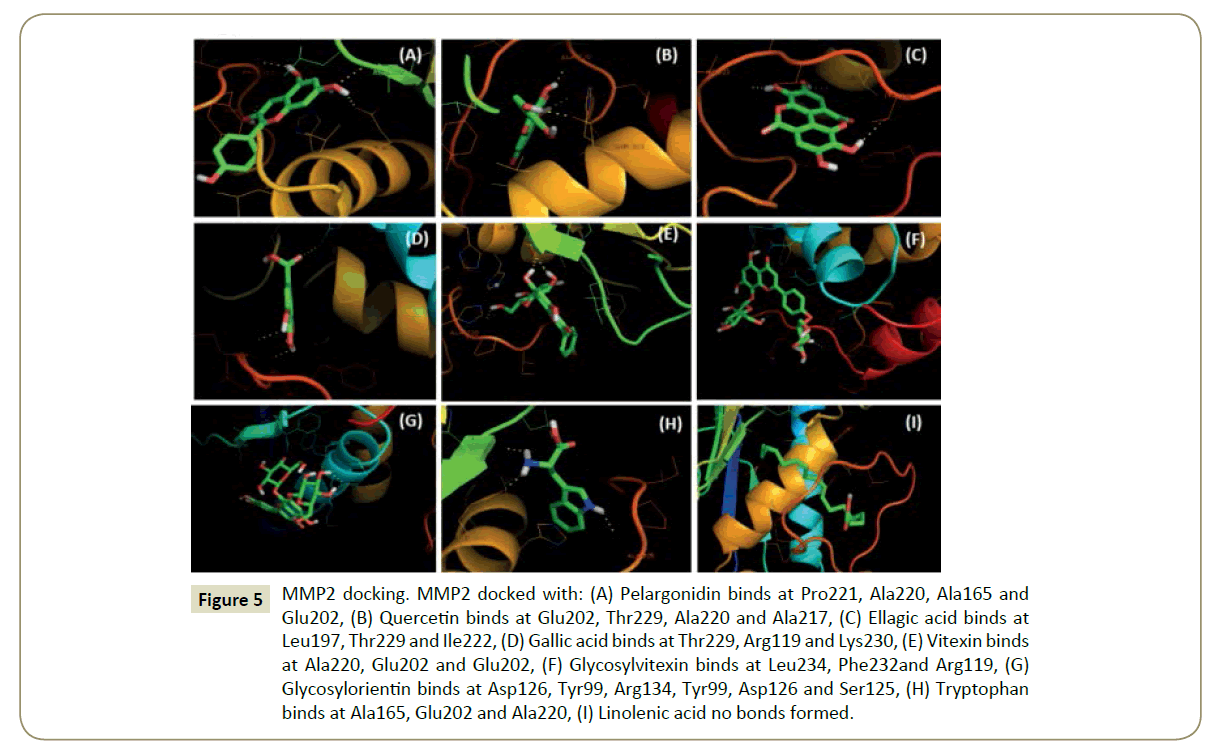
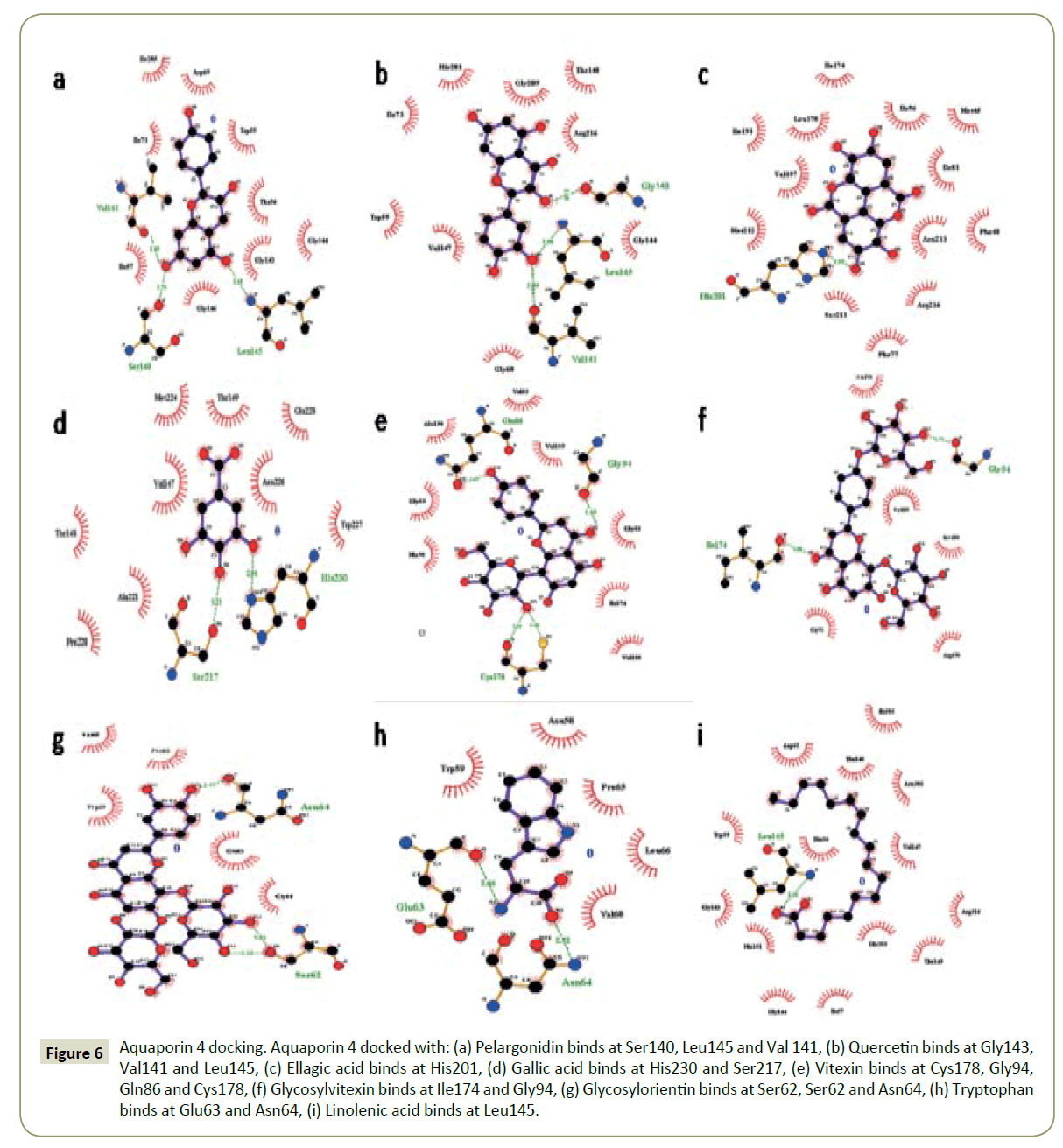
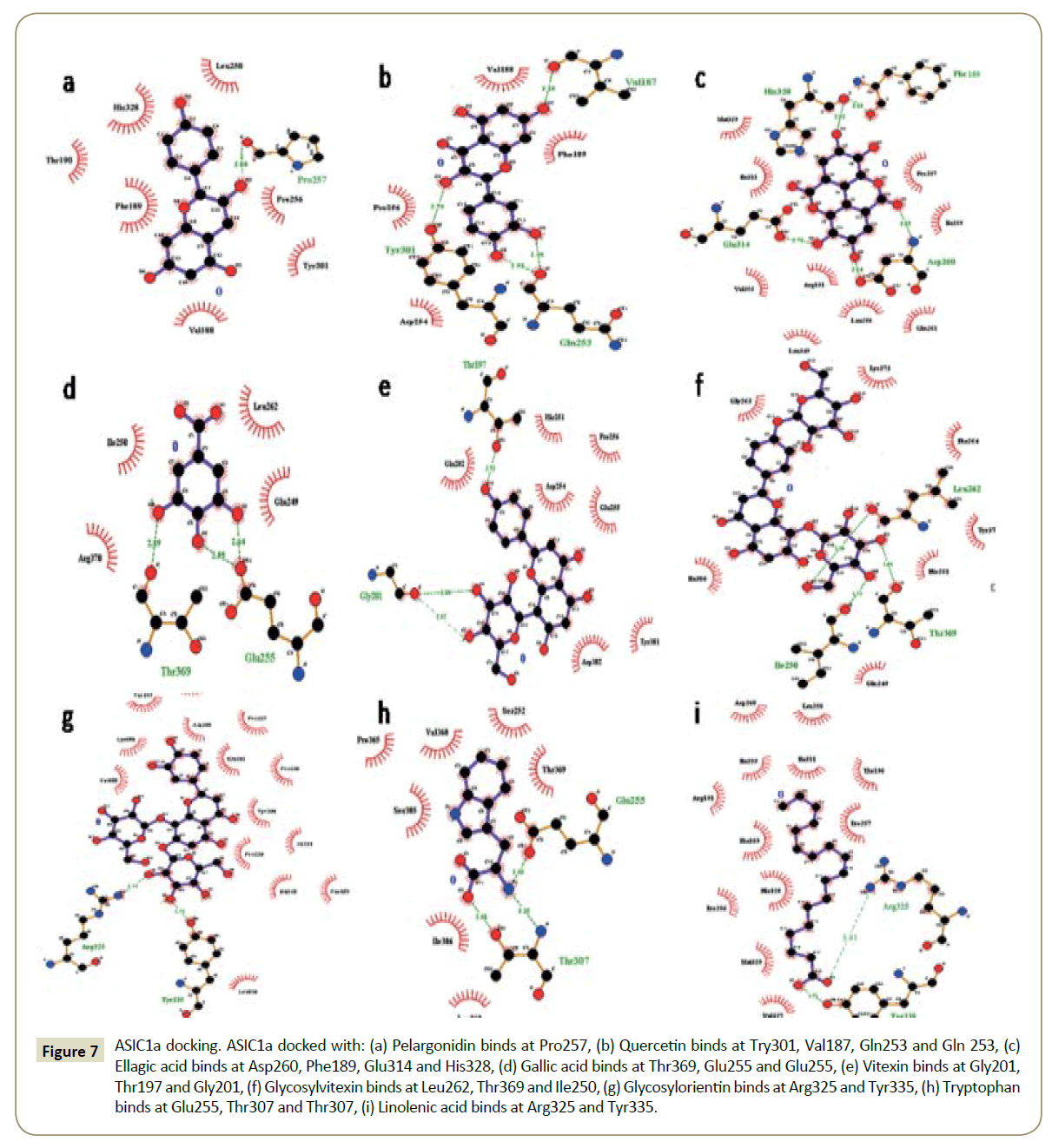
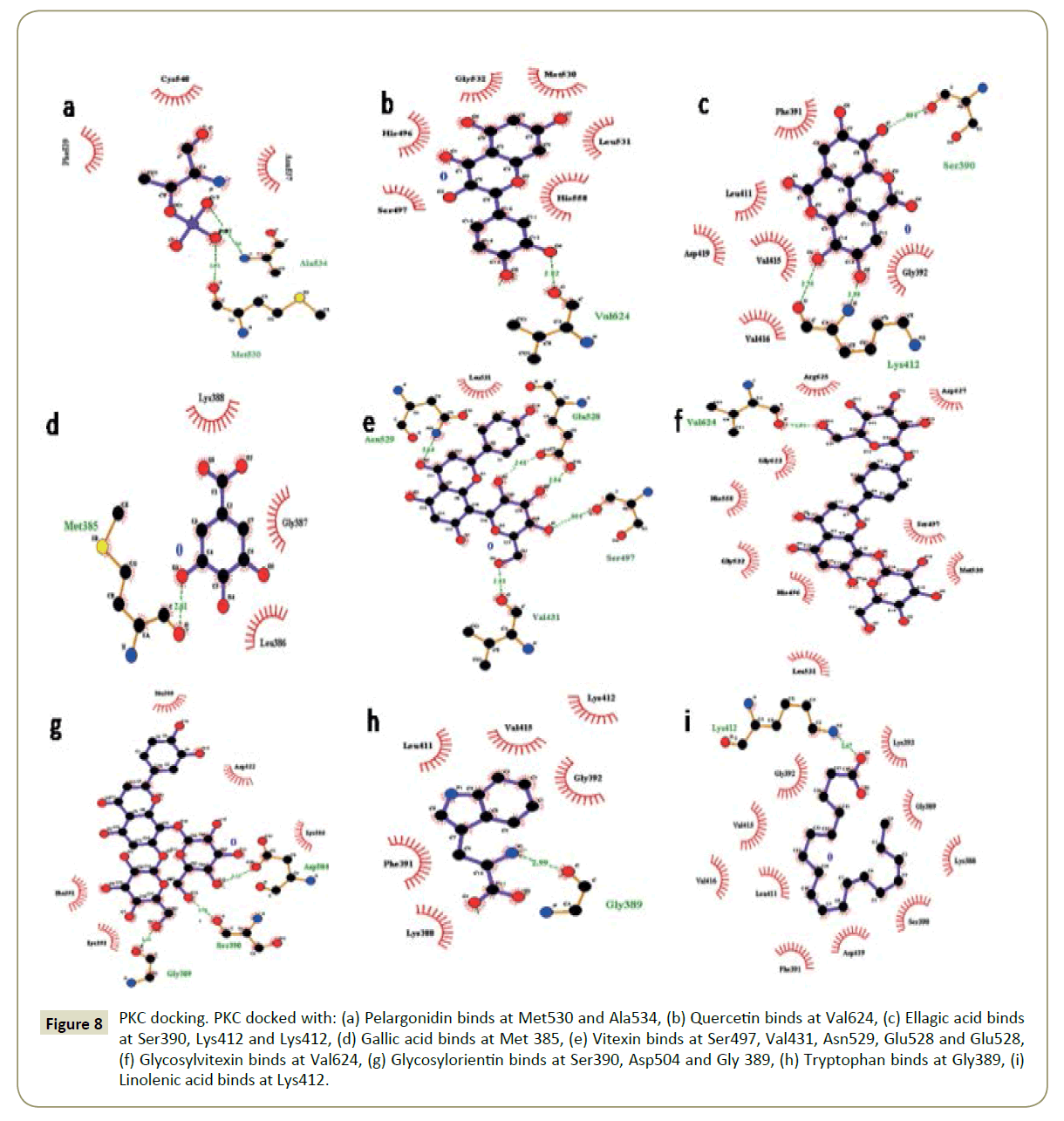
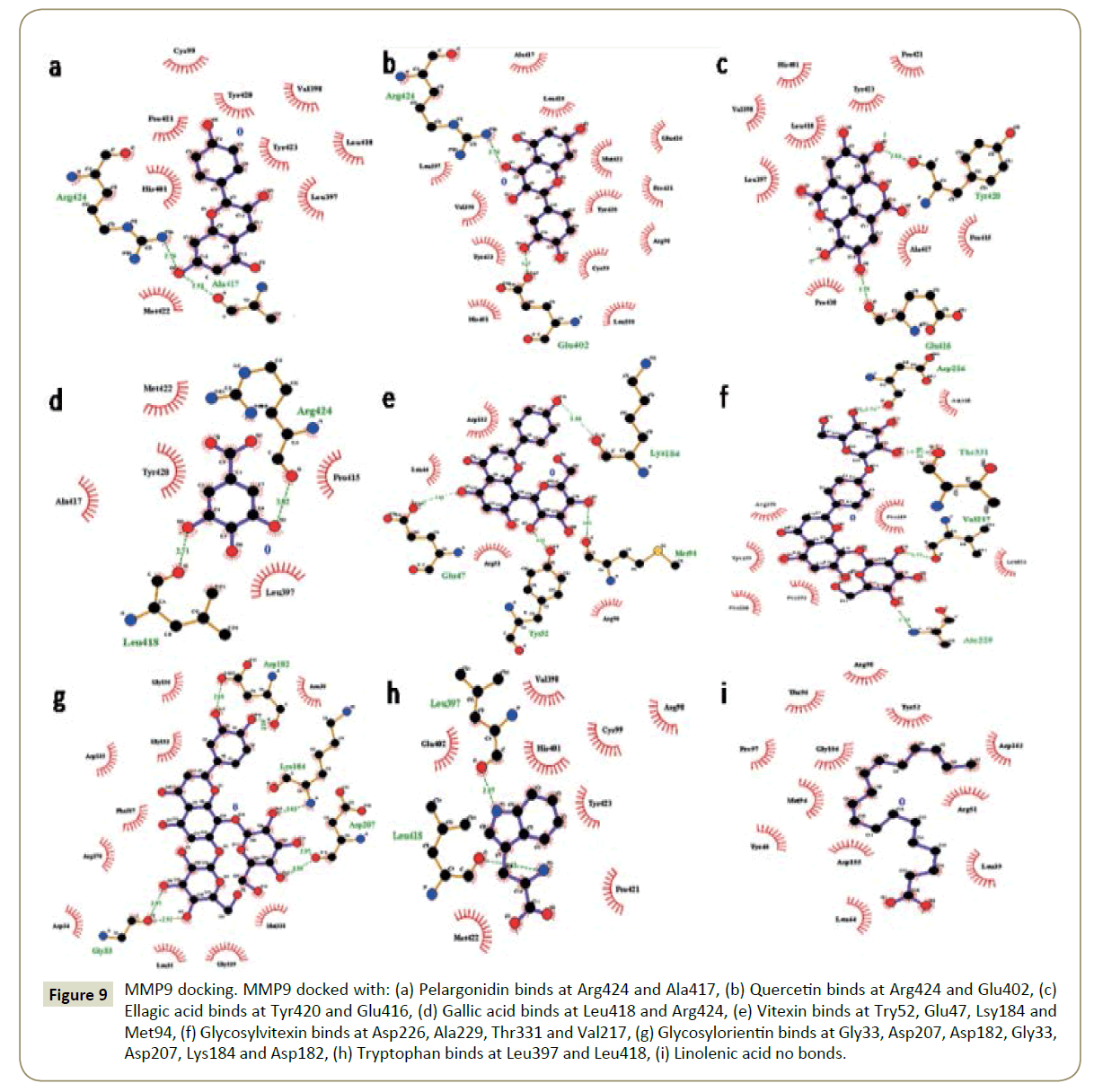
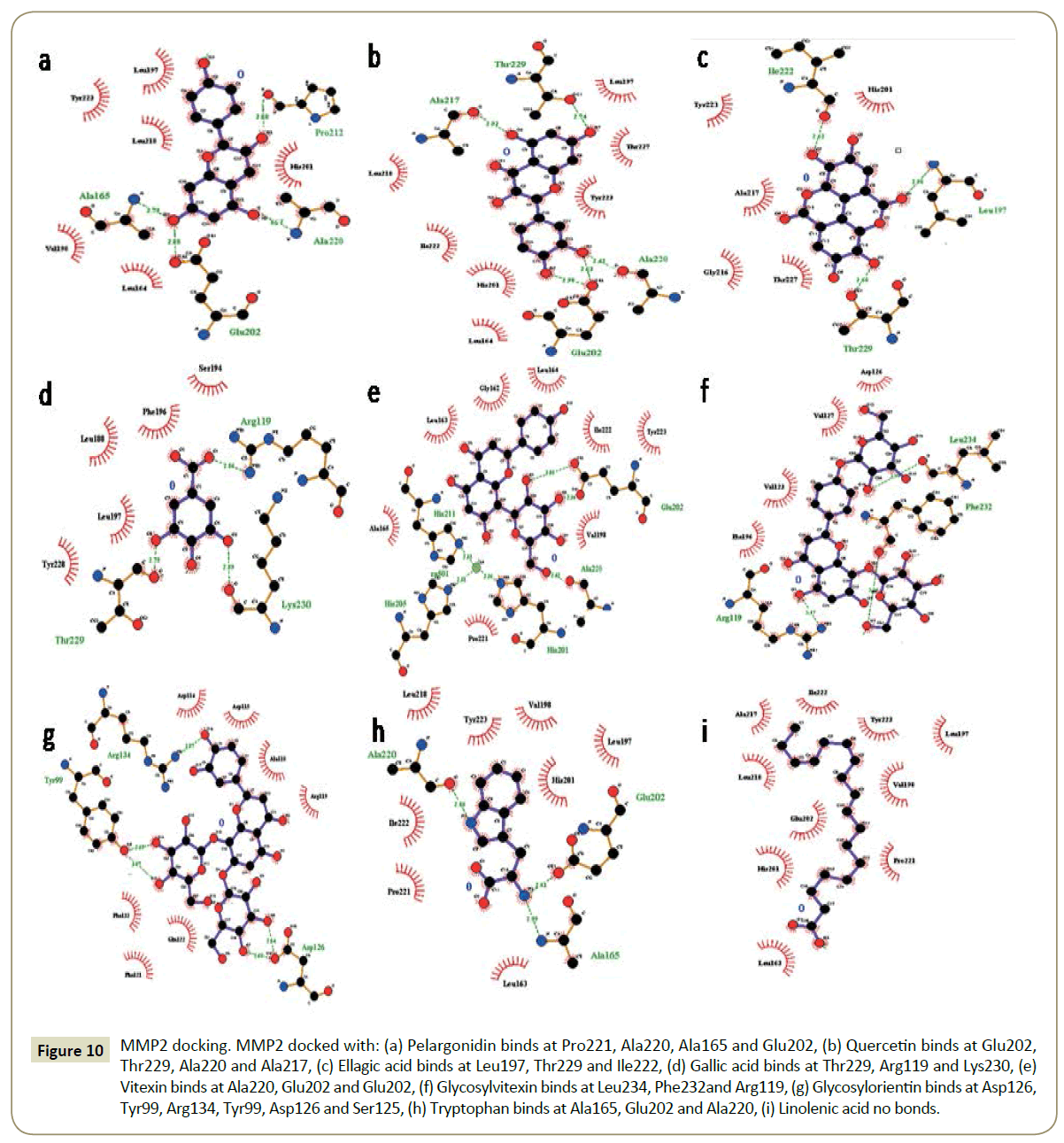
The binding pockets of pelargonidin in MMP9 (Figures 1-10) consist of amino acids Arg424 and Ala417 (Figures 4A and 9a). The binding pocket for MMP2 showed 4 H-bonds with Pro212, Ala220, Ala165 and Glu202 thus is more effective than that of MMP9 (Figures 5A and 10a). Aquaporin4 docking active site residues are Ser 140, Leu145 and Val 141 (Figures 1A and 6a). Pelargonidin and ASIC1a binding occurs at Pro257 (Figures 2A and 7a). The docking with Protein Kinase C resulted in 2 H-bonds at residues Met530 and Ala534 (Figures 3A and 8a).
Figure 1: Aquaporin 4 docking. Aquaporin 4 docked with: (A) Pelargonidin binds at Ser140, Leu145 and Val 141, (B) Quercetin binds at Gly143, Val141 and Leu145, (C) Ellagic acid binds at His201, (D) Gallic acid binds at His230 and Ser217, (E) Vitexin binds at Cys178, Gly94, Gln86 and Cys178, (F) Glycosylvitexin binds at Ile174 and Gly94, (G) Glycosylorientin binds at Ser62, Ser62 and Asn64, (H) Tryptophan binds at Glu63 and Asn64, (I) Linolenic acid binds at Leu145.
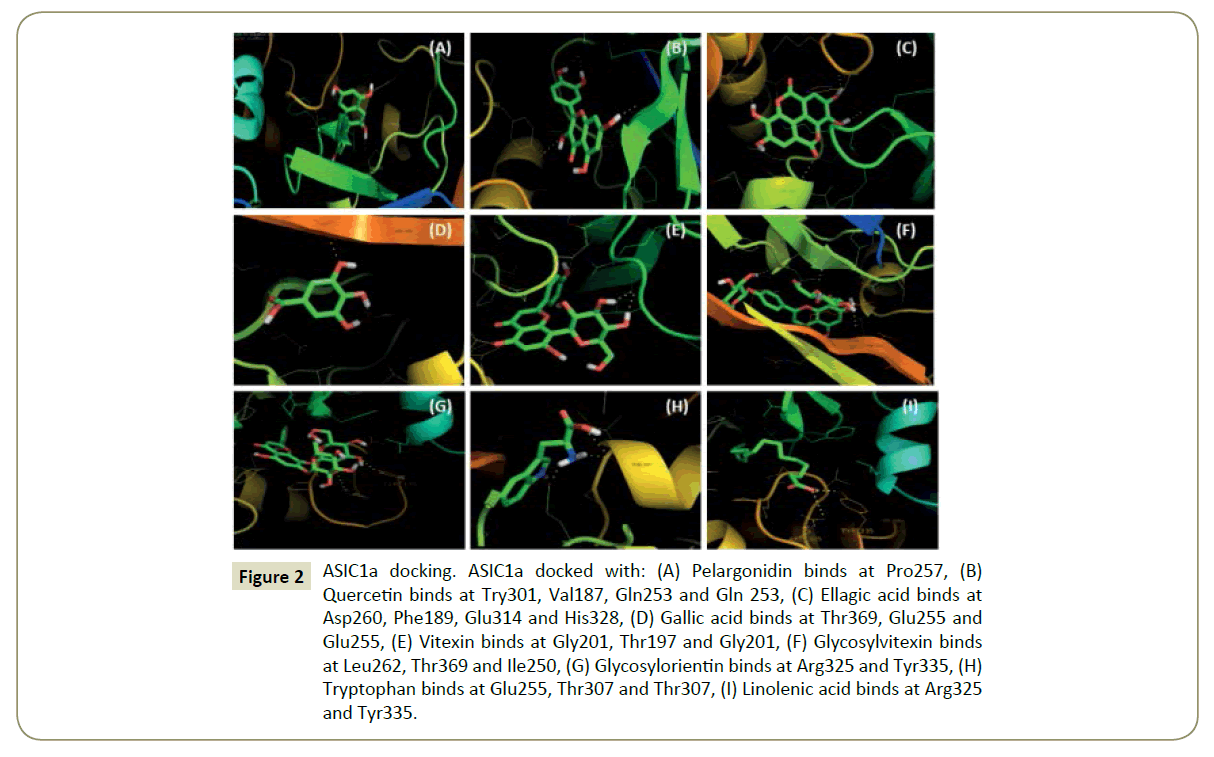
Figure 2: ASIC1a docking. ASIC1a docked with: (A) Pelargonidin binds at Pro257, (B) Quercetin binds at Try301, Val187, Gln253 and Gln 253, (C) Ellagic acid binds at Asp260, Phe189, Glu314 and His328, (D) Gallic acid binds at Thr369, Glu255 and Glu255, (E) Vitexin binds at Gly201, Thr197 and Gly201, (F) Glycosylvitexin binds at Leu262, Thr369 and Ile250, (G) Glycosylorientin binds at Arg325 and Tyr335, (H) Tryptophan binds at Glu255, Thr307 and Thr307, (I) Linolenic acid binds at Arg325 and Tyr335.
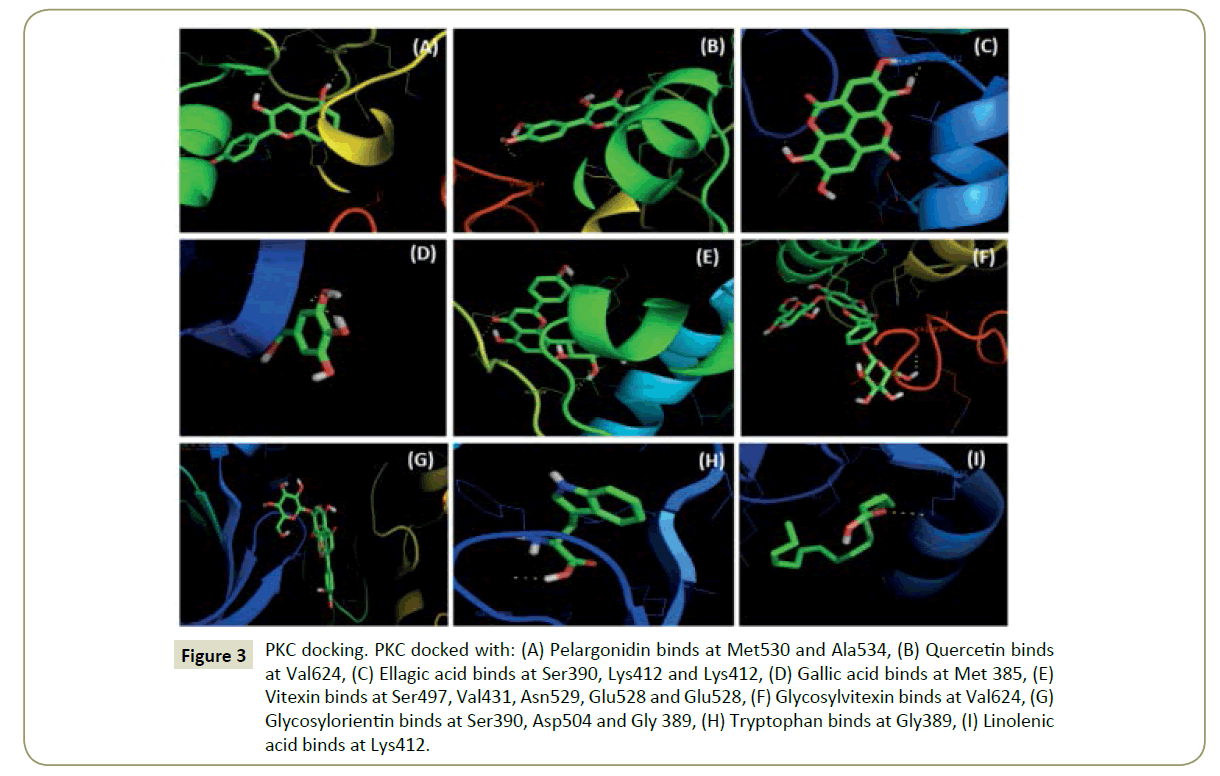
Figure 3: PKC docking. PKC docked with: (A) Pelargonidin binds at Met530 and Ala534, (B) Quercetin binds at Val624, (C) Ellagic acid binds at Ser390, Lys412 and Lys412, (D) Gallic acid binds at Met 385, (E) Vitexin binds at Ser497, Val431, Asn529, Glu528 and Glu528, (F) Glycosylvitexin binds at Val624, (G) Glycosylorientin binds at Ser390, Asp504 and Gly 389, (H) Tryptophan binds at Gly389, (I) Linolenic acid binds at Lys412.
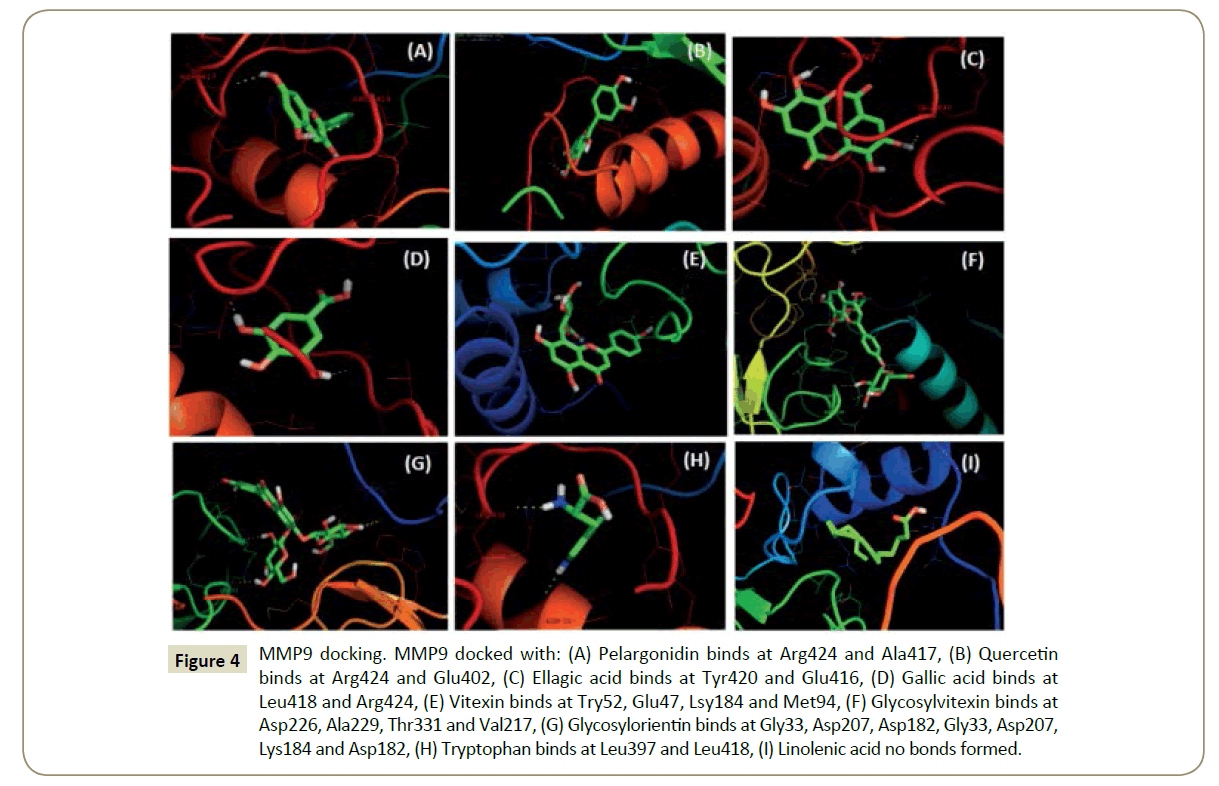
Figure 4: MMP9 docking. MMP9 docked with: (A) Pelargonidin binds at Arg424 and Ala417, (B) Quercetin binds at Arg424 and Glu402, (C) Ellagic acid binds at Tyr420 and Glu416, (D) Gallic acid binds at Leu418 and Arg424, (E) Vitexin binds at Try52, Glu47, Lsy184 and Met94, (F) Glycosylvitexin binds at Asp226, Ala229, Thr331 and Val217, (G) Glycosylorientin binds at Gly33, Asp207, Asp182, Gly33, Asp207, Lys184 and Asp182, (H) Tryptophan binds at Leu397 and Leu418, (I) Linolenic acid no bonds formed.
Figure 5: MMP2 docking. MMP2 docked with: (A) Pelargonidin binds at Pro221, Ala220, Ala165 and Glu202, (B) Quercetin binds at Glu202, Thr229, Ala220 and Ala217, (C) Ellagic acid binds at Leu197, Thr229 and Ile222, (D) Gallic acid binds at Thr229, Arg119 and Lys230, (E) Vitexin binds at Ala220, Glu202 and Glu202, (F) Glycosylvitexin binds at Leu234, Phe232and Arg119, (G) Glycosylorientin binds at Asp126, Tyr99, Arg134, Tyr99, Asp126 and Ser125, (H) Tryptophan binds at Ala165, Glu202 and Ala220, (I) Linolenic acid no bonds formed.
Figure 6: Aquaporin 4 docking. Aquaporin 4 docked with: (a) Pelargonidin binds at Ser140, Leu145 and Val 141, (b) Quercetin binds at Gly143, Val141 and Leu145, (c) Ellagic acid binds at His201, (d) Gallic acid binds at His230 and Ser217, (e) Vitexin binds at Cys178, Gly94, Gln86 and Cys178, (f) Glycosylvitexin binds at Ile174 and Gly94, (g) Glycosylorientin binds at Ser62, Ser62 and Asn64, (h) Tryptophan binds at Glu63 and Asn64, (i) Linolenic acid binds at Leu145.
Figure 7: ASIC1a docking. ASIC1a docked with: (a) Pelargonidin binds at Pro257, (b) Quercetin binds at Try301, Val187, Gln253 and Gln 253, (c) Ellagic acid binds at Asp260, Phe189, Glu314 and His328, (d) Gallic acid binds at Thr369, Glu255 and Glu255, (e) Vitexin binds at Gly201, Thr197 and Gly201, (f) Glycosylvitexin binds at Leu262, Thr369 and Ile250, (g) Glycosylorientin binds at Arg325 and Tyr335, (h) Tryptophan binds at Glu255, Thr307 and Thr307, (i) Linolenic acid binds at Arg325 and Tyr335.
Figure 8: PKC docking. PKC docked with: (a) Pelargonidin binds at Met530 and Ala534, (b) Quercetin binds at Val624, (c) Ellagic acid binds at Ser390, Lys412 and Lys412, (d) Gallic acid binds at Met 385, (e) Vitexin binds at Ser497, Val431, Asn529, Glu528 and Glu528, (f) Glycosylvitexin binds at Val624, (g) Glycosylorientin binds at Ser390, Asp504 and Gly 389, (h) Tryptophan binds at Gly389, (i) Linolenic acid binds at Lys412.
Figure 9: MMP9 docking. MMP9 docked with: (a) Pelargonidin binds at Arg424 and Ala417, (b) Quercetin binds at Arg424 and Glu402, (c) Ellagic acid binds at Tyr420 and Glu416, (d) Gallic acid binds at Leu418 and Arg424, (e) Vitexin binds at Try52, Glu47, Lsy184 and Met94, (f) Glycosylvitexin binds at Asp226, Ala229, Thr331 and Val217, (g) Glycosylorientin binds at Gly33, Asp207, Asp182, Gly33, Asp207, Lys184 and Asp182, (h) Tryptophan binds at Leu397 and Leu418, (i) Linolenic acid no bonds.
Figure 10: MMP2 docking. MMP2 docked with: (a) Pelargonidin binds at Pro221, Ala220, Ala165 and Glu202, (b) Quercetin binds at Glu202, Thr229, Ala220 and Ala217, (c) Ellagic acid binds at Leu197, Thr229 and Ile222, (d) Gallic acid binds at Thr229, Arg119 and Lys230, (e) Vitexin binds at Ala220, Glu202 and Glu202, (f) Glycosylvitexin binds at Leu234, Phe232and Arg119, (g) Glycosylorientin binds at Asp126, Tyr99, Arg134, Tyr99, Asp126 and Ser125, (h) Tryptophan binds at Ala165, Glu202 and Ala220, (i) Linolenic acid no bonds.
Quercetin
The docking study of quercetin with MMP9, MMP2, Aquaporin 4, ASIC1a, and PKC stabilized the target proteins with the binding energies of -10.41,-9.32,-7.57,-6.4 and -6.38 kcal/mol respectively (Table 2). Docking of quercetin resulted in binding with MMP9 at Arg424 and Glu402 with 2 H-bonds (Figures 4B and 9b). Binding conformations for MMP2 have residues Glu202, Thr229, Ala 220 and Ala217 with 4 H-bonds (Figures 5B and 10b). Active site pocket for Aquaporin 4 were found to have residues Gly143, Val141 and Leu145 (Figures 1B and 6b). The binding of quercetin with ASIC1a is also potent having 4 H-bond at residues Try301, Val187, Gln253 and Gln 253 (Figures 2B and 7b). The binding from docking with Protein Kinase C resulted at residue Val624 (Figures 3B and 8b).
Ellagic acid
Docking of ellagic acid with MMP9, MMP2, Aquaporin 4, ASIC1a, and PKC stabilize the target proteins with the binding energies of -8.46, -7.56, -3.22, -9.93, and -7.11 kcal/mol respectively (Table 2). With MMP9 it gave 2 H-bonds with residues Tyr420 and Glu416 (Figures 4C and 9c). MMP2 binding resulted at residues Leu197, Thr229 and Ile222 (Figures 5C and 10c). ASIC1a binding site residues are Asp260, Phe189, Glu314 and His328 (Figures 2C and 7c) which advocates its potential against ASIC1a. Binding with Aquaporin 4 is at His 201 residue (Figures 1C and 6c). Active site in Protein Kinase C resulted at Ser390, Lys412 and Lys412 residues (Figures 3C and 8c).
Gallic acid
Gallic acid blind docking with MMP9, MMP2, Aquaporin 4, ASIC1a, and PKC stabilized the target proteins with the binding energies of -4.27, -4.44, -4.19, -4.46 and -4.1 kcal/mol respectively (Table 2). Gallic acid inhibited MMP9 by binding with residues Leu418 and Arg424 (Figures 4D and 9d). MMP2 binding took place at Thr229, Arg119 and Lys230 (Figures 5D and 10d). Aquaporin 4 docking resulted in active site at residues His230 and Ser217 (Figures 1D and 6d). When docked with ASIC1a the active site residues were found to be Thr369, Glu255 and Glu255 (Figures 2D and 7d). Protein Kinase C inhibition efficacy of gallic acid established with 2 H-bonds at residue Met 385 (Figures 3D and 8d).
Vitexin
Vitexin docking stabilized targets MMP9, MMP2, Aquaporin 4, ASIC1a, and PKC with the binding energies of -7.71, -8.63, -6.01, -7.07 and -4.97 kcal/mol respectively (Table 2). MMP9 docking resulted in catalytic site residues Try52, Glu47, Lsy184 and Met94 (Figures 4E and 9e). Docking of MMP2 consists of binding pocket residues Ala220, Glu202 and Glu202 (Figures 5E and 10e). ASIC1a with vitexin bonded at Gly201, Thr197 and Gly201 active site residues (Figures 2E and 7e). Aquaporin 4 binding residues were Cys178, Gly94, Gln86 and Cys178 (Figures 1E and 6e). Binding with Protein Kinase C provided Ser497, Val431, Asn529, Glu528 and Glu528 as active site residues (Figures 3E and 8e).
Glycosylvitexin
Glycosylvitexin docking with MMP9, MMP2, Aquaporin 4, ASIC1a, and PKC stabilized the target proteins with the binding energies of -5.51, -6.29, -5.07, -4.63 and -6.11 kcal/mol respectively (Table 2). The docking of glycosylvitexin with MMP9 resulted with Asp226, Ala229, Thr331 and Val217 forming its binding pocket (Figures 4F and 9f). MMP2 active binding site had residues Leu234, Phe232 and Arg119 (Figures 5F and 10f). Aquaporin 4 docking gave binding pocket site residues Ile174 and Gly94 (Figures 1F, and 6f). Docking with ASIC1a resulted in 3 H-bonds with residues Leu262, Thr369 and Ile250 (Figures 2F and 7f). Protein Kinase C formed 1H bond at residue Val624 (Figures 3F and 8f).
Glycosylorientin
The docking of glycosylorientin with MMP9, MMP2, Aquaporin 4, ASIC1a, and PKC stabilised the target proteins with the binding energies of -6.15, -4.15, -3.51, -5.13 and -4.62 kcal/mol respectively (Table 2). Glycosylorientin with MMP9 gave an outstanding result with residues Gly33, Asp207, Asp182, Gly33, Asp207, Lys184 and Asp182 (Figures 4G and 9g). MMP2 binding occurred at residues Asp126, Thr99, Arg134, Tyr99, and Asp126 (Figures 5G and 10g). With Aquaporin 4 binding site residues are Ser62, Ser62 and Asn64 (Figures 1G and 6g). When docked with ASIC1a it gave residue Arg325 and Tyr335 as catalytic site residues (Figures 2G and 7g). Docking of glycosylorientin with Protein Kinase C provided binding pocket at residues Ser390, Asp504 and Gly 389 (Figures 3G and 8g).
Tryptophan
The docking study of tryptophan with MMP9, MMP2, Aquaporin 4, ASIC1a, and PKC stabilised the target proteins with the binding energies of -7.86, -8.31, -5.84, -6.62 and -5.58kcal/mol respectively (Table 2). MMP9 binding pocket residues were Leu397 and Leu418 (Figures 4H and 9h). MMP2 active site residues were Ala165, Glu202, and Ala220 (Figures 5H and 10h). The docking with Aquaporin 4 gave Glu63 and Asn64 in binding pocket (Figures 1H and 6h). In ASIC1a, tryptophan formed bonds with Glu255, Thr307 and Thr307 residues (Figures 2H and 7h). With PKC the active site residue was Gly389 (Figures 3H and 8h).
Linolenic acid
Linolenic acid stabilized MMP9, MMP2, Aquaporin 4, ASIC1a, and PKC with the binding energies of -5.42, -6.24, -4.51, -5.64 and -4.3 kcal/mol respectively. With ASIC1a, Arg325 and Tyr335 binding pocket residues resulted (Figures 2I and 7i). Aquaporin 4 active site residue was Leu 415 having 2 H-bonds (Figures 1I and 6i). Docking with PKC gave binding at Lys412 residue (Figures 3I and 8i) whereas no hydrogen bonds formed with MMP2 and MMP9 (Figures 4I, 5I, 9i, and 10i).
Discussion
Pearl millet is a cheap crop usually used for fodder and feed. It is consumed by rural society as well as the urban society of the developed and developing countries. It has great nutritional value along with some components with ultimate effective properties which can prove them a cure for most widespread neurodegenerative disorders [2]. These components are Pelargonidin, Quercetin, Ellagic acid, Gallic acid, Vitexin, Glycosylvitexin, Glycosylorientin, Tryptophan and Linolenic acid which have anti-oxidant and anti-inflammatory properties [5]. As now a day’s neurodegenerative diseases are becoming a very common cause of death, it needs to be intervened and new efficacious treatment is demanded. The treatments available till date are not effective to target the cause efficiently due to unknown targets and ambiguous pathology in some cases and also in their inability to pass the blood brain barrier. Neurodegenerative disorders like cerebral ischemia, Alzheimer’s disease, Parkinson’s disease, multiple sclerosis etc. are yelling at millions of victims and need to be treated effectively and at early stages [9]. To fulfil the purpose of developing an effective treatment for these diseases we analysed constituents of pearl millet having antioxidant and anti-inflammatory property by molecular docking with the potent targets of various neurodegenerative diseases. According to pathological analysis of these diseases we found MMP9, MMP2, Aquaporin 4, ASIC1a and PKC playing great roles in progression of these diseases [11,13] It has been well documented that acidosis, activation of matrix metalloproteinases and cerebral edema are the main pathological feature which occurs due to dysfunction of ASIC1a channels, MMP9 and MMP2 enzymes and Aquaporin 4 respectively in cerebral ischemia [11]. Through our docking analysis, we found Pelargonidin, quercetin, vitexin, glycosylvitexin, glycosylorientin, ellagic acid and gallic acid, constituents of pearl millet binding effectively and inhibiting the ASIC1a, MMP9, MMP2 and Aquaporin 4 targets of cerebral ischemia proving their potency as effective treatments for cerebral ischemia. It has been reported that synaptic plasticity and memory loss due to inter and intracellular accumulation of beta amyloid protein and neurofibrillary tangles is a main pathological feature in Alzheimer’s disease. Regulation of levels of MMP2 and MMP9 and role of PKC was found to be responsible for these pathologies respectively. So, in the case of Alzheimer’s also the constituents of pearl millet proved themselves effective by binding with high binding energies with MMP2, MMP9 and also with PKC [7]. Vitexin showed the best result in the context of PKC by forming 5 H-bonds with the molecule. As neurofibrillary tangles and amyloid plaque are also involved in pathology of Parkinson’s disease and blocks transmission through cortex region or perforant tracks from areas of association of isocortex to hippocampus and cause memory impairment thus same targets as that of Alzheimer’s can work for the treatment of the disease by inhibition by the above mentioned constituents of pearl millet [19]. In multiple sclerosis, MMP9 and ASIC1a elevated levels cause havoc by increasing the ion concentrations and early relapse which can be targeted and inhibited effectively by glycosylorientin, glycosylvetixin and vitexin (for MMP) and pelargonidin and quercetin (for ASIC1a), proving their efficiency to cure this disease [18]. So, our hypothesis of using nutraceutical property of pearl millet as a potential treatment for the majority of neurodegenerative disorders is having a great scope as suggested by our docking results. Thus, incorporation of pearl millet in diet can give good effects for improving the diseased conditions and curing them. The economically cheap and high nutraceutical value of the constituents of pearl millets demands for increased [20,21] consumption and research is needed to invent ways to exploit its nutritional efficiency to maximum.
Conclusion
In accordance with our docking studies, we found that the use of pearl millet as a dietary supplement has a great potential to treat the majority of NDs, due to its antioxidant and anti-inflammatory properties of its constituents along with their good efficacy to inhibit potent molecular targets of these diseases. So, increased intake of pearl millet in diet can give unbelievable results for curing the NDs as well as other diseases caused due to oxidative stress. On the basis of our findings and the previous reports, we can conclude that more research is required, to increase and enhance such properties of pearl millet by various genetic modification techniques and also for developing efficient natural drugs compounds for treating specific diseases for the purpose of reducing the risk of neurodegenerative diseases in rural and the urban areas of the developed as well as developing countries.
Conflict of Interest
The authors declare no conflict in interests.
Funding
We have received no funding for this work.
References
- Kannappan R, Gupta SC, Kim JH, Reuter S, Aggarwal BB (2011) Neuroprotection by spice-derived nutraceuticals: you are what you eat! Mol Neurobiol 44: 142-159.
- Pushparaj FS, Urooj A (2014) Antioxidant activity in two pearl millet (Pennisetumtyphoideum) cultivars as influenced by processing. Antioxidants 3: 55-66.
- Kaur KD, Jha A, Sabikhi L, Singh AK (2014) Significance of coarse cereals in health and nutrition: a review. J Food Sci Technol 51: 1429-1441.
- NambiarVanisha S, Dhaduk J J, Sareen N, Shahu T, Desai R (2011) Potential functional implications of pearl millet (Pennisetum glaucum) in health and disease. J Appl Pharmaceut Sci 1: 62-67.
- Ali K, Khalid D, Marmouzi I (2015) In vitro antioxidant and anti-inflammatory activities of extracts of pearl millet (Pennisetum glaucum L.). J Food Sci Technol 7: 1-6.
- Mouquet-Rivier C, Icard-Vernière C, Guyot JP, Tou Eh, Rochette I (2008) Consumption pattern, biochemical composition and nutritional value of fermented pearl millet gruels in Burkina Faso. Int J Food Sci Nutr 59: 716-729.
- Hjorth E, Zhu M, Toro VC, Inger V, Jan P, et al. (2013) Omega-3 fatty acids enhance phagocytosis of Alzheimer's disease-related amyloid-beta42 by human microglia and decrease inflammatory markers. J Alzheimers Dis 35: 697-713.
- Gupta V, Nagar R (2010) Effect of cooking, fermentation, dehulling and utensils on antioxidants present in pearl millet rabadi-a traditional fermented food. J Food Sci Technol 47: 73-76.
- Gabathuler R (2010) Approaches to transport therapeutic drugs across the blood-brain barrier to treat brain diseases. Neurobiol Dis 37: 48-57.
- Lobo V, Patil A, Phatak A, Chandra N (2010) Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn Rev 4: 118-126.
- Pandey AK, Verma S, Bhattacharya P, Paul S, Mishra A, et al. (2012) An In-Silico strategy to explore neuroprotection by quercetin in cerebral ischemia: A novel hypothesis based on inhibition of matrix metalloproteinase (MMPs) and acid sensing ion channel 1a (ASIC1a). Medical Hypotheses 79: 76-81.
- Bhattacharya P, Pandey AK, Shukla SC (2013) Neuroprotection by μ-calpain and matrix metalloproteinases inhibition by Piroxicam in cerebral ischemia: an in silico study. Med Chem Res 22: 5112-5119.
- Bhattacharya P, Pandey AK, Paul S (2012) Aquaporin-4 in cerebral edema following ischemia/reperfusion injury: exploration of novel therapeutic strategies. Am J Neuroprot Neuroregen 4: 1-27.
- Lorenzl S, Albers DS, Relkin N, Ngyuen T, Hilgenberg SL, et al. (2003) Increased plasma levels of matrix metalloproteinase-9 in patients with Alzheimer's disease. Neurochem Int 43: 191-196.
- Lim NK, Villemagne VL, Soon CP, Laughton KM, Rowe CC, et al. (2011) Investigation of matrix metalloproteinases, MMP-2 and MMP-9, in plasma reveals a decrease of MMP-2 in Alzheimer's disease. J Alzheimers Dis 26: 779-786.
- Lan Y L, Zhao J, Ma T, Li S (2016) The potential roles of aquaporin 4 in Alzheimer's disease. Mol Neurobiol 53: 5300-5309.
- Avolio C, Ruggieri M, Giuliani F, Liuzzi GM, Leante R, et al. (2003) Serum MMP-2 and MMP-9 are elevated in different multiple sclerosis subtypes. J Neuroimmunol 136: 46-53.
- Arun T, Tomassini V, Sbardella E, De Ruiter MB, Matthews L, et al. (2013) Targeting ASIC1 in primary progressive multiple sclerosis: evidence of neuroprotection with amiloride. Brain 136: 106-115.
- Chu XP, Xiong ZG (2012) Physiological and pathological functions of acid-sensing ion channels in the central nervous system. Curr Drug Targets 13: 263-271.
- Pandey KB, Rizvi SI (2009) Plant polyphenols as dietary antioxidants in human health and disease. Oxid Med Cell Longev 2: 270-278.
- Gandhi S, Abramov AY (2012) Mechanism of Oxidative Stress in Neurodegeneration. Oxid Med Cell Longev 2012: 11.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences